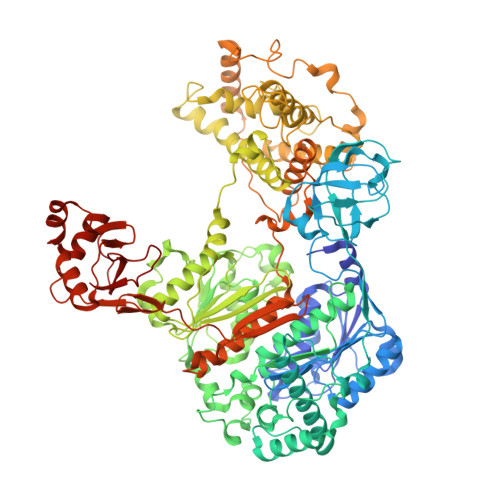
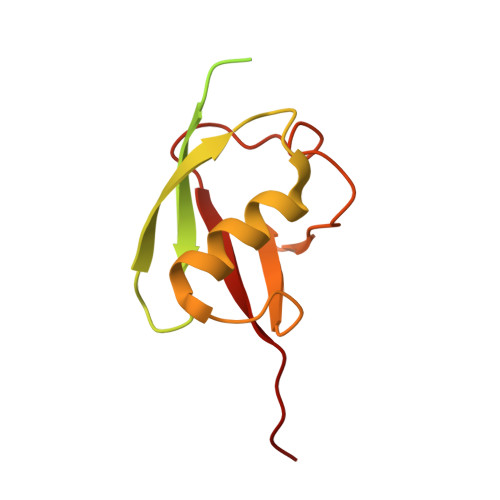
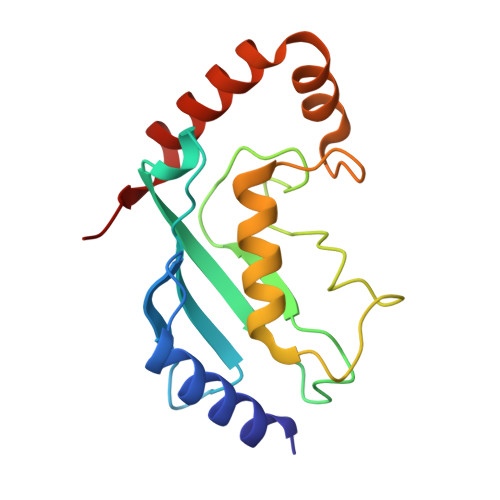
Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Afsar, M., Liu, G., Jia, L., Ruben, E.A., Nayak, D., Sayyad, Z., Bury, P.D.S., Cano, K.E., Nayak, A., Zhao, X.R., Shukla, A., Sung, P., Wasmuth, E.V., Gack, M.U., Olsen, S.K.(2023) Nat Commun 14: 4786-4786
- PubMed: 37553340
- DOI: https://doi.org/10.1038/s41467-023-39780-z
- Primary Citation of Related Structures:
8SE9, 8SEA, 8SEB, 8SV8 - PubMed Abstract:
ISG15 plays a crucial role in the innate immune response and has been well-studied due to its antiviral activity and regulation of signal transduction, apoptosis, and autophagy. ISG15 is a ubiquitin-like protein that is activated by an E1 enzyme (Uba7) and transferred to a cognate E2 enzyme (UBE2L6) to form a UBE2L6-ISG15 intermediate that functions with E3 ligases that catalyze conjugation of ISG15 to target proteins. Despite its biological importance, the molecular basis by which Uba7 catalyzes ISG15 activation and transfer to UBE2L6 is unknown as there is no available structure of Uba7. Here, we present cryo-EM structures of human Uba7 in complex with UBE2L6, ISG15 adenylate, and ISG15 thioester intermediate that are poised for catalysis of Uba7-UBE2L6-ISG15 thioester transfer. Our structures reveal a unique overall architecture of the complex compared to structures from the ubiquitin conjugation pathway, particularly with respect to the location of ISG15 thioester intermediate. Our structures also illuminate the molecular basis for Uba7 activities and for its exquisite specificity for ISG15 and UBE2L6. Altogether, our structural, biochemical, and human cell-based data provide significant insights into the functions of Uba7, UBE2L6, and ISG15 in cells.
- Department of Biochemistry & Structural Biology, University of Texas Health Science Center at San Antonio, San Antonio, TX, 78229, USA.
Organizational Affiliation: