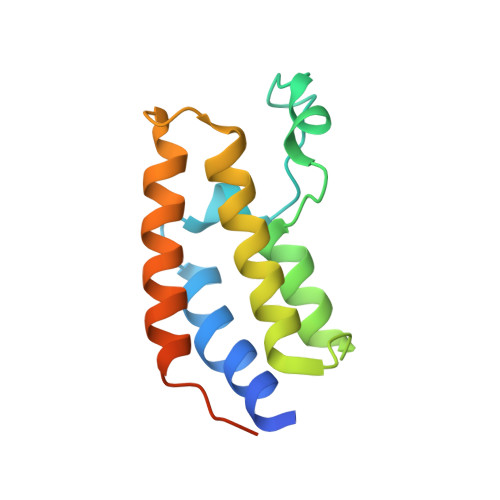
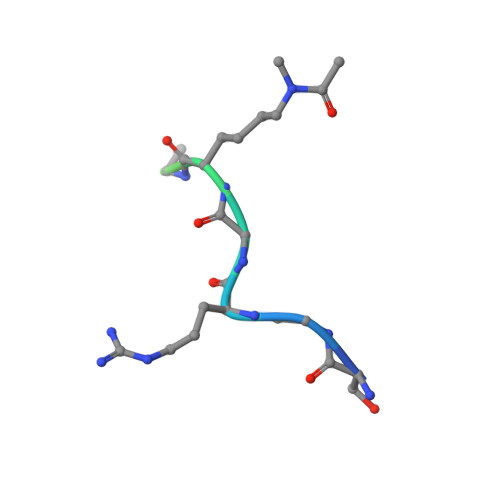
Acetyl-methyllysine marks chromatin at active transcription start sites.
Lu-Culligan, W.J., Connor, L.J., Xie, Y., Ekundayo, B.E., Rose, B.T., Machyna, M., Pintado-Urbanc, A.P., Zimmer, J.T., Vock, I.W., Bhanu, N.V., King, M.C., Garcia, B.A., Bleichert, F., Simon, M.D.(2023) Nature 622: 173-179
- PubMed: 37731000
- DOI: https://doi.org/10.1038/s41586-023-06565-9
- Primary Citation of Related Structures:
8SB6 - PubMed Abstract:
Lysine residues in histones and other proteins can be modified by post-translational modifications that encode regulatory information 1 . Lysine acetylation and methylation are especially important for regulating chromatin and gene expression 2-4 . Pathways involving these post-translational modifications are targets for clinically approved therapeutics to treat human diseases. Lysine methylation and acetylation are generally assumed to be mutually exclusive at the same residue. Here we report cellular lysine residues that are both methylated and acetylated on the same side chain to form N ε -acetyl-N ε -methyllysine (Kacme). We show that Kacme is found on histone H4 (H4Kacme) across a range of species and across mammalian tissues. Kacme is associated with marks of active chromatin, increased transcriptional initiation and is regulated in response to biological signals. H4Kacme can be installed by enzymatic acetylation of monomethyllysine peptides and is resistant to deacetylation by some HDACs in vitro. Kacme can be bound by chromatin proteins that recognize modified lysine residues, as we demonstrate with the crystal structure of acetyllysine-binding protein BRD2 bound to a histone H4Kacme peptide. These results establish Kacme as a cellular post-translational modification with the potential to encode information distinct from methylation and acetylation alone and demonstrate that Kacme has all the hallmarks of a post-translational modification with fundamental importance to chromatin biology.
- Department of Molecular Biophysics & Biochemistry, Yale University, New Haven, CT, USA.
Organizational Affiliation: