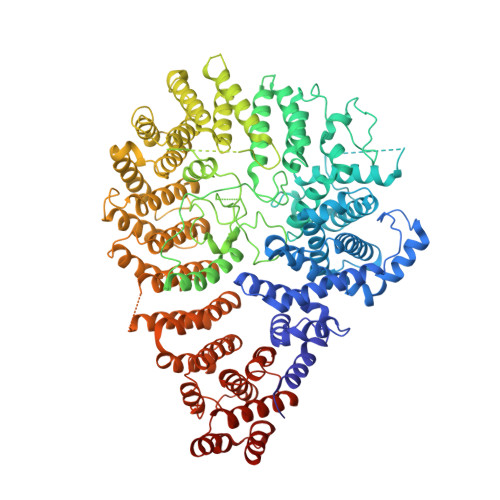
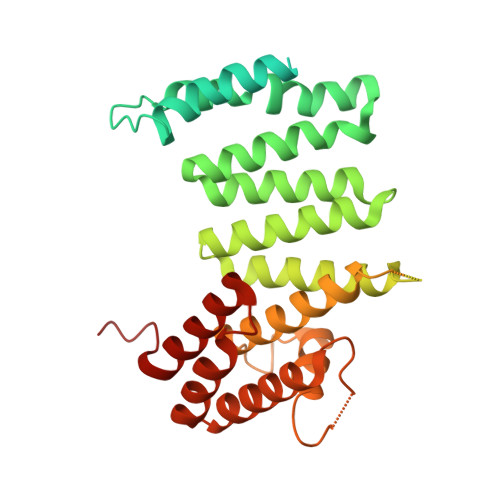
Delineation of functional subdomains of Huntingtin protein and their interaction with HAP40.
Alteen, M.G., Deme, J.C., Alvarez, C.P., Loppnau, P., Hutchinson, A., Seitova, A., Chandrasekaran, R., Silva Ramos, E., Secker, C., Alqazzaz, M., Wanker, E.E., Lea, S.M., Arrowsmith, C.H., Harding, R.J.(2023) Structure 31: 1121-1131.e6
- PubMed: 37390814
- DOI: https://doi.org/10.1016/j.str.2023.06.002
- Primary Citation of Related Structures:
8SAH - PubMed Abstract:
The huntingtin (HTT) protein plays critical roles in numerous cellular pathways by functioning as a scaffold for its many interaction partners and HTT knock out is embryonic lethal. Interrogation of HTT function is complicated by the large size of this protein so we studied a suite of structure-rationalized subdomains to investigate the structure-function relationships within the HTT-HAP40 complex. Protein samples derived from the subdomain constructs were validated using biophysical methods and cryo-electron microscopy, revealing they are natively folded and can complex with validated binding partner, HAP40. Derivatized versions of these constructs enable protein-protein interaction assays in vitro, with biotin tags, and in cells, with luciferase two-hybrid assay-based tags, which we use in proof-of-principle analyses to further interrogate the HTT-HAP40 interaction. These open-source biochemical tools enable studies of fundamental HTT biochemistry and biology, will aid the discovery of macromolecular or small-molecule binding partners and help map interaction sites across this large protein.
- Structural Genomics Consortium, University of Toronto, Toronto, ON M5G 1L7, Canada; POINT Biopharma, 22 St Clair Avenue E Suite 1201, Toronto, ON M4T 2S3, Canada.
Organizational Affiliation: