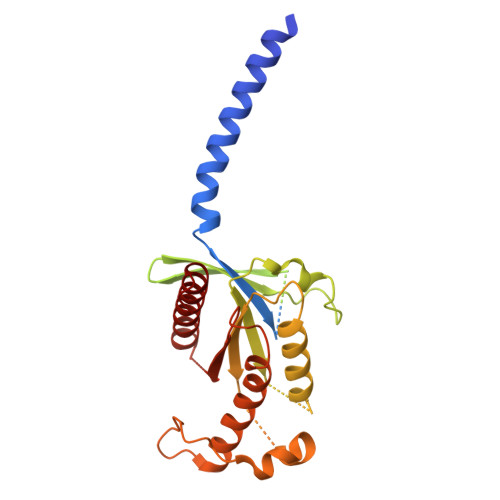
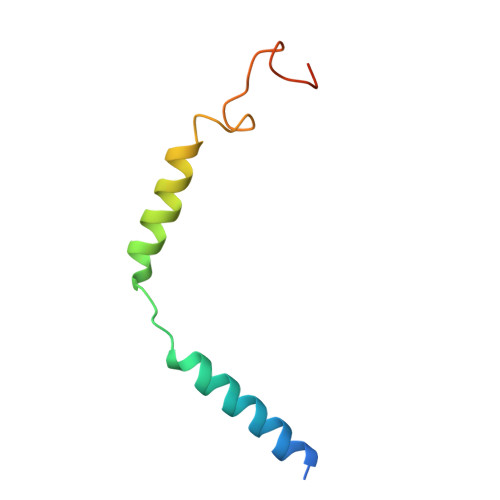
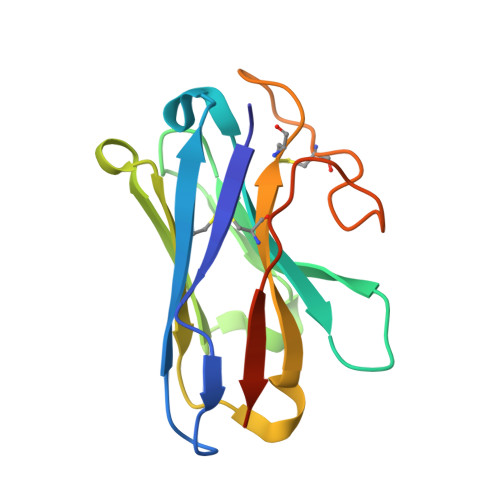
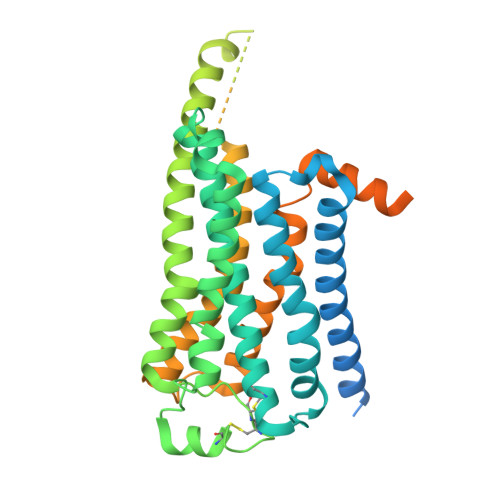
Cryo-EM structure of a cell-free synthesized full-length human beta 1-adrenergic receptor in complex with G s .
Merino, F., Kock, Z., Ermel, U., Dahlhaus, P., Grimm, A., Seybert, A., Kubicek, J., Frangakis, A.S., Dotsch, V., Hilger, D., Bernhard, F.(2025) Structure 33: 1867
- PubMed: 40858117
- DOI: https://doi.org/10.1016/j.str.2025.07.020
- Primary Citation of Related Structures:
8S2T - PubMed Abstract:
The third intracellular loop (ICL3) of the β1-adrenergic receptor (β1AR) plays a critical role in regulating G protein coupling, yet the structural basis has remained unclear due to truncations of ICL3 in all available structures of the β1AR in complex with G s or a G protein mimetic nanobody. To address this, we used cell-free cotranslational insertion of full-length human β1AR into nanodiscs and determined its cryo-electron microscopy (cryo-EM) structure in complex with G s . In this structure, ICL3 extends transmembrane helix 5, resulting in enhanced interactions with Gα s and in a slight rotation of the engaged G protein. This repositioning enables new polar interactions between Gα s , ICL2 and helix 8, while ICL1 and helix 8 form additional contacts with Gβ. These structural insights, supported by mutational analysis, demonstrate that ICL3 enhances G protein activation and downstream cAMP signaling by promoting more extensive interactions between the receptor and the heterotrimeric G protein.
- Cube Biotech GmbH, Monheim, Germany.
Organizational Affiliation: