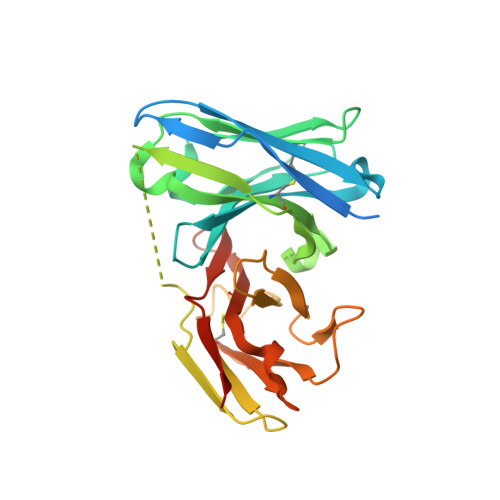
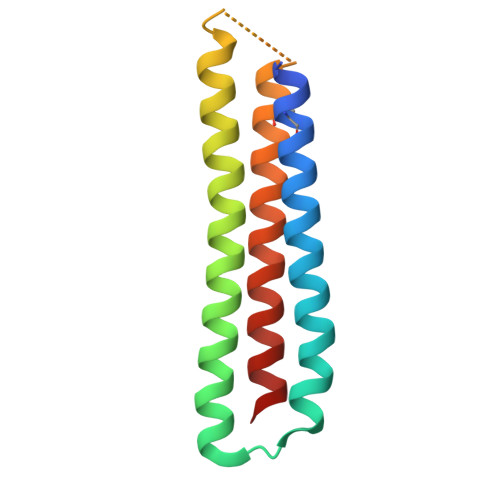
Rational structure-guided design of a blood stage malaria vaccine immunogen presenting a single epitope from PfRH5.
Harrison, T.E., Alam, N., Farrell, B., Quinkert, D., Lias, A.M., King, L.D.W., Barfod, L.K., Draper, S.J., Campeotto, I., Higgins, M.K.(2024) EMBO Mol Med 16: 2539-2559
- PubMed: 39223355
- DOI: https://doi.org/10.1038/s44321-024-00123-0
- Primary Citation of Related Structures:
8RZ0, 8RZ1, 8RZ2 - PubMed Abstract:
There is an urgent need for improved malaria vaccine immunogens. Invasion of erythrocytes by Plasmodium falciparum is essential for its life cycle, preceding symptoms of disease and parasite transmission. Antibodies which target PfRH5 are highly effective at preventing erythrocyte invasion and the most potent growth-inhibitory antibodies bind a single epitope. Here we use structure-guided approaches to design a small synthetic immunogen, RH5-34EM which recapitulates this epitope. Structural biology and biophysics demonstrate that RH5-34EM is correctly folded and binds neutralising monoclonal antibodies with nanomolar affinity. In immunised rats, RH5-34EM induces PfRH5-targeting antibodies that inhibit parasite growth. While PfRH5-specific antibodies were induced at a lower concentration by RH5-34EM than by PfRH5, RH5-34EM induced antibodies that were a thousand-fold more growth-inhibitory as a factor of PfRH5-specific antibody concentration. Finally, we show that priming with RH5-34EM and boosting with PfRH5 achieves the best balance between antibody quality and quantity and induces the most effective growth-inhibitory response. This rationally designed vaccine immunogen is now available for use as part of future malaria vaccines, alone or in combination with other immunogens.
- Department of Biochemistry, Dorothy Crowfoot Hodgkin Building, University of Oxford, South Parks Rd, Oxford, OX1 3QU, UK.
Organizational Affiliation: