Structural characterization of the ACDC domain from ApiAP2 proteins, a potential molecular target against apicomplexan parasites.
Le Berre, M., Tubiana, T., Reutersward Waldner, P., Lazar, N., Li de la Sierra-Gallay, I., Santos, J.M., Llinas, M., Nessler, S.(2025) Acta Crystallogr D Struct Biol 81: 38-48
- PubMed: 39820027
- DOI: https://doi.org/10.1107/S2059798324012518
- Primary Citation of Related Structures:
8RWU, 8RXA, 8RXO - PubMed Abstract:
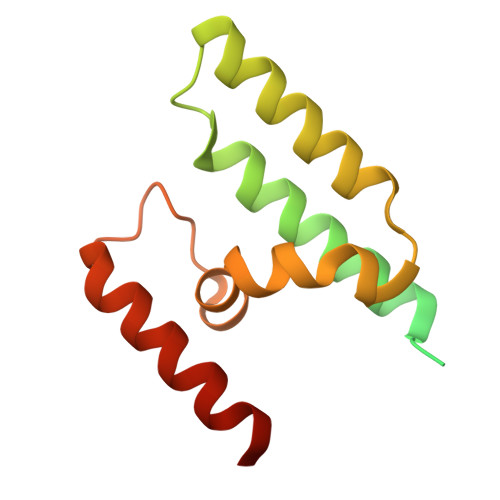
The apicomplexan AP2 (ApiAP2) proteins are the best characterized family of DNA-binding proteins in Plasmodium spp. malaria parasites. Apart from the AP2 DNA-binding domain, there is little sequence similarity between ApiAP2 proteins. However, a conserved AP2-coincident domain mostly at the C-terminus (ACDC domain) is observed in a subset of the ApiAP2 proteins. The structure and function of this domain remain unknown. We report two crystal structures of ACDC domains derived from distinct Plasmodium ApiAP2 proteins, revealing a conserved, unique, noncanonical, four-helix bundle architecture. We used these structures to perform in silico docking calculations against a library of known antimalarial compounds and identified potential small-molecule ligands that bind in a highly conserved hydrophobic pocket that is present in all apicomplexan ACDC domains. These ligands provide a new molecular basis for the future design of ACDC inhibitors.
- Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 91198 Gif-sur-Yvette, France.
Organizational Affiliation: