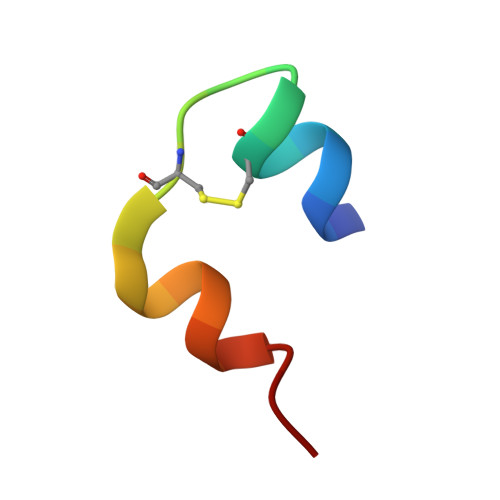
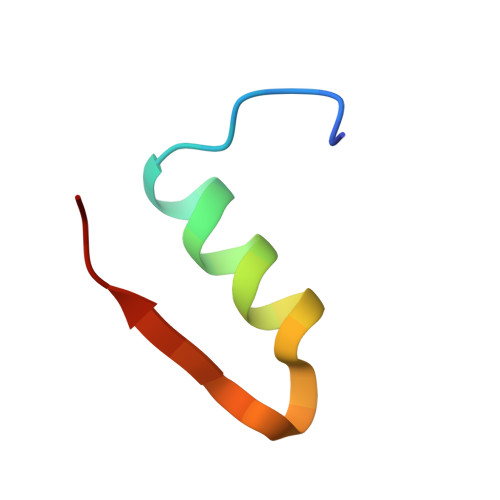
Enhanced disulphide bond stability contributes to the once-weekly profile of insulin icodec.
Hubalek, F., Cramer, C.N., Helleberg, H., Johansson, E., Nishimura, E., Schluckebier, G., Steensgaard, D.B., Sturis, J., Kjeldsen, T.B.(2024) Nat Commun 15: 6124-6124
- PubMed: 39033137
- DOI: https://doi.org/10.1038/s41467-024-50477-9
- Primary Citation of Related Structures:
8RRP - PubMed Abstract:
Insulin icodec is a once-weekly insulin analogue that has a long half-life of approximately 7 days, making it suitable for once weekly dosing. The Insulin icodec molecule was developed based on the hypothesis that lowering insulin receptor affinity and introducing a strong albumin-binding moiety would result in a long insulin half-life, provided that non-receptor-mediated clearance is diminished. Here, we report an insulin clearance mechanism, resulting in the splitting of insulin molecules into its A-chain and B-chain by a thiol-disulphide exchange reaction. Even though the substitutions in insulin icodec significantly stabilise insulin against such degradation, some free B-chain is observed in plasma samples from minipigs and people with type 2 diabetes. In summary, we identify thiol-disulphide exchange reactions to be an important insulin clearance mechanism and find that stabilising insulin icodec towards this reaction significantly contributes to its long pharmacokinetic/pharmacodynamic profile.
- Novo Nordisk A/S, Maaloev, Denmark. FHUB@novonordisk.com.
Organizational Affiliation: