Structural basis for regulation of a CBASS-CRISPR-Cas defense island by a transmembrane anti-sigma factor and its ECF sigma partner.
Bernal-Bernal, D., Pantoja-Uceda, D., Lopez-Alonso, J.P., Lopez-Rojo, A., Lopez-Ruiz, J.A., Galbis-Martinez, M., Ochoa-Lizarralde, B., Tascon, I., Elias-Arnanz, M., Ubarretxena-Belandia, I., Padmanabhan, S.(2024) Sci Adv 10: eadp1053-eadp1053
- PubMed: 39454004
- DOI: https://doi.org/10.1126/sciadv.adp1053
- Primary Citation of Related Structures:
8RLZ, 8ROT, 8RW5 - PubMed Abstract:
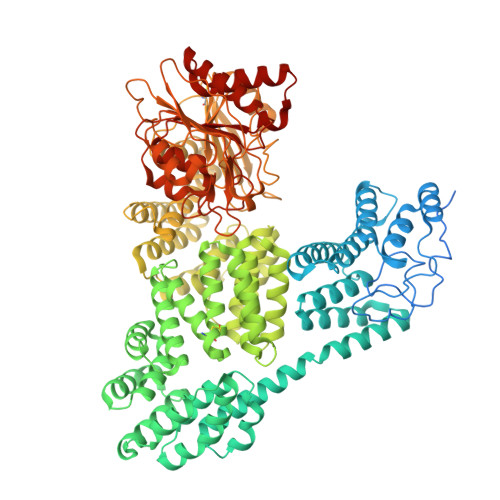
How CRISPR-Cas and cyclic oligonucleotide-based antiphage signaling systems (CBASS) are coordinately deployed against invaders remains unclear. We show that a locus containing two CBASS and one type III-B CRISPR-Cas system, regulated by the transmembrane anti-σ DdvA and its cognate extracytoplasmic function (ECF) σ DdvS, can defend Myxococcus xanthus against a phage. Cryo-electron microscopy reveals DdvA-DdvS pairs assemble as arrow-shaped transmembrane dimers. Each DdvA periplasmic domain adopts a separase/craspase-type tetratricopeptide repeat (TPR)-caspase HetF-associated with TPR (TPR-CHAT) architecture with an incomplete His-Cys active site, lacking three α-helices conserved among CHAT domains. Each active site faces the dimer interface, raising the possibility that signal-induced caspase-like DdvA autoproteolysis in trans precedes RseP-mediated intramembrane proteolysis and DdvS release. Nuclear magnetic resonance reveals a DdvA cytoplasmic CHCC-type zinc-bound three-helix bundle that binds to DdvS σ 2 and σ 4 domains, undergoing σ 4 -induced helix extension to trap DdvS. Altogether, we provide structural-mechanistic insights into membrane anti-σ-ECF σ regulation of an antiviral CBASS-CRISPR-Cas defense island.
- Departamento de Genética y Microbiología, Área de Genética (Unidad Asociada al IQF-CSIC), Universidad de Murcia, 30100 Murcia, Spain.
Organizational Affiliation: