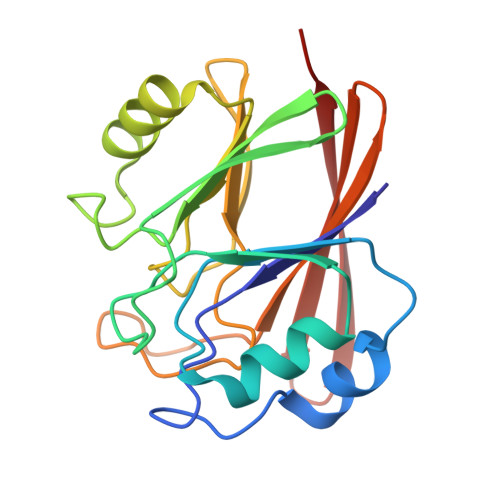
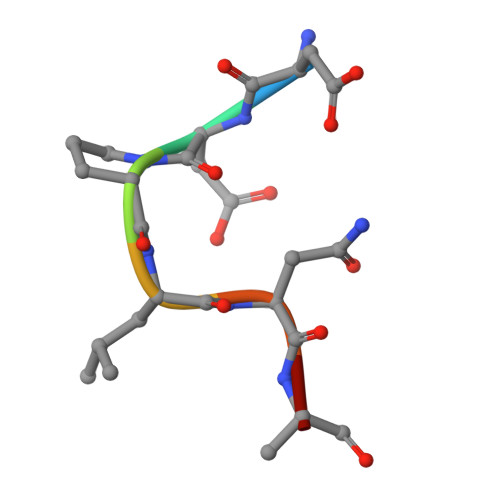
Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Romano-Moreno, M., Astorga-Simon, E.N., Rojas, A.L., Hierro, A.(2024) Protein Sci 33: e4980-e4980
- PubMed: 38607248
- DOI: https://doi.org/10.1002/pro.4980
- Primary Citation of Related Structures:
8RKS - PubMed Abstract:
Endosomal trafficking ensures the proper distribution of lipids and proteins to various cellular compartments, facilitating intracellular communication, nutrient transport, waste disposal, and the maintenance of cell structure. Retromer, a peripheral membrane protein complex, plays an important role in this process by recruiting the associated actin-polymerizing WASH complex to establish distinct sorting domains. The WASH complex is recruited through the interaction of the VPS35 subunit of retromer with the WASH complex subunit FAM21. Here, we report the identification of two separate fragments of FAM21 that interact with VPS35, along with a third fragment that binds to the VPS29 subunit of retromer. The crystal structure of VPS29 bound to a peptide derived from FAM21 shows a distinctive sharp bend that inserts into a conserved hydrophobic pocket with a binding mode similar to that adopted by other VPS29 effectors. Interestingly, despite the network of interactions between FAM21 and retromer occurring near the Parkinson's disease-linked mutation (D620N) in VPS35, this mutation does not significantly impair the direct association with FAM21 in vitro.
- Center for Cooperative Research in Biosciences (CIC bioGUNE), Bilbao, Spain.
Organizational Affiliation: