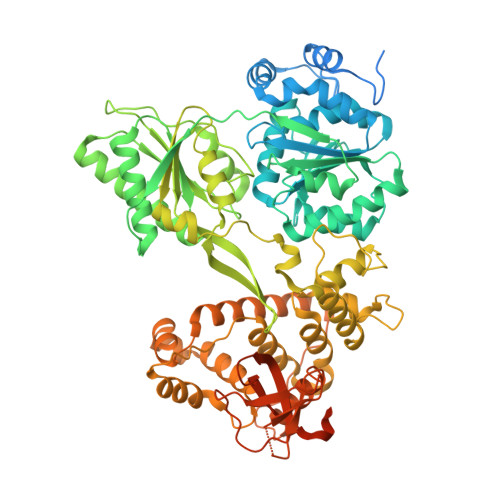
An inhibitory segment within G-patch activators tunes Prp43-ATPase activity during ribosome assembly.
Portugal-Calisto, D., Geiger, A.G., Rabl, J., Vadas, O., Oborska-Oplova, M., Mazur, J., Richina, F., Klingauf-Nerurkar, P., Michel, E., Leitner, A., Boehringer, D., Panse, V.G.(2024) Nat Commun 15: 10150-10150
- PubMed: 39578461
- DOI: https://doi.org/10.1038/s41467-024-54584-5
- Primary Citation of Related Structures:
8RDY - PubMed Abstract:
Mechanisms by which G-patch activators tune the processive multi-tasking ATP-dependent RNA helicase Prp43 (DHX15 in humans) to productively remodel diverse RNA:protein complexes remain elusive. Here, a comparative study between a herein and previously characterized activators, Tma23 and Pxr1, respectively, defines segments that organize Prp43 function during ribosome assembly. In addition to the activating G-patch, we discover an inhibitory segment within Tma23 and Pxr1, I-patch, that restrains Prp43 ATPase activity. Cryo-electron microscopy and hydrogen-deuterium exchange mass spectrometry show how I-patch binds to the catalytic RecA-like domains to allosterically inhibit Prp43 ATPase activity. Tma23 and Pxr1 contain dimerization segments that organize Prp43 into higher-order complexes. We posit that Prp43 function at discrete locations on pre-ribosomal RNA is coordinated through toggling interactions with G-patch and I-patch segments. This could guarantee measured and timely Prp43 activation, enabling precise control over multiple RNA remodelling events occurring concurrently during ribosome formation.
- Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.
Organizational Affiliation: