Trypanosoma brucei Invariant Surface Glycoprotein 75 Is an Immunoglobulin Fc Receptor Inhibiting Complement Activation and Antibody-Mediated Cellular Phagocytosis.
Mikkelsen, J.H., Stodkilde, K., Jensen, M.P., Hansen, A.G., Wu, Q., Lorentzen, J., Graversen, J.H., Andersen, G.R., Fenton, R.A., Etzerodt, A., Thiel, S., Andersen, C.B.F.(2024) J Immunol 212: 1334-1344
- PubMed: 38391367
- DOI: https://doi.org/10.4049/jimmunol.2300862
- Primary Citation of Related Structures:
8RD2 - PubMed Abstract:
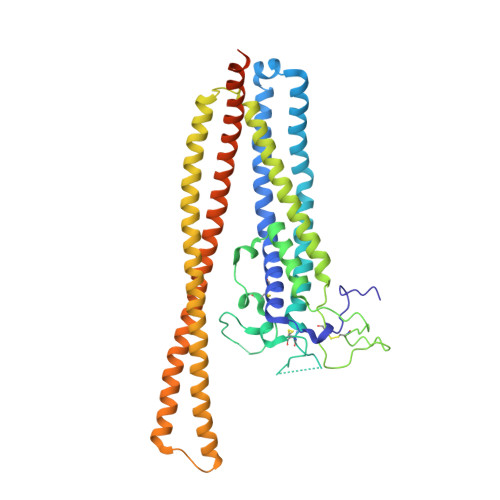
Various subspecies of the unicellular parasite Trypanosoma brucei cause sleeping sickness, a neglected tropical disease affecting millions of individuals and domestic animals. Immune evasion mechanisms play a pivotal role in parasite survival within the host and enable the parasite to establish a chronic infection. In particular, the rapid switching of variant surface glycoproteins covering a large proportion of the parasite's surface enables the parasite to avoid clearance by the adaptive immune system of the host. In this article, we present the crystal structure and discover an immune-evasive function of the extracellular region of the T. brucei invariant surface gp75 (ISG75). Structural analysis determined that the ISG75 ectodomain is organized as a globular head domain and a long slender coiled-coil domain. Subsequent ligand screening and binding analysis determined that the head domain of ISG75 confers interaction with the Fc region of all subclasses of human IgG. Importantly, the ISG75-IgG interaction strongly inhibits both activation of the classical complement pathway and Ab-dependent cellular phagocytosis by competing with C1q and host cell FcγR CD32. Our data reveal a novel immune evasion mechanism of T. brucei, with ISG75 able to inactivate the activities of Abs recognizing the parasite surface proteins.
- Department of Biomedicine, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: