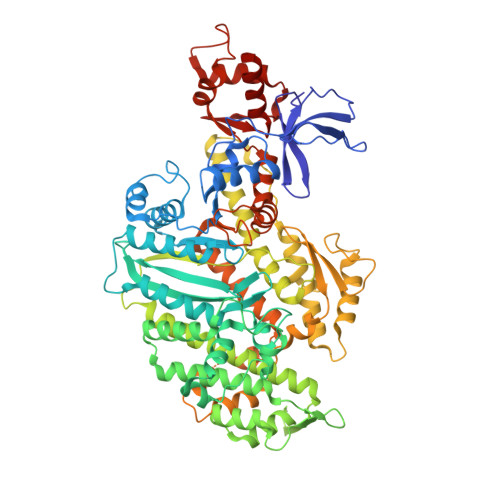
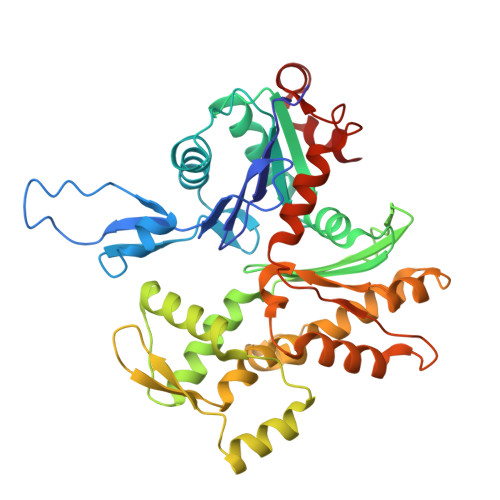
Swinging lever mechanism of myosin directly shown by time-resolved cryo-EM.
Klebl, D.P., McMillan, S.N., Risi, C., Forgacs, E., Virok, B., Atherton, J.L., Harris, S.A., Stofella, M., Winkelmann, D.A., Sobott, F., Galkin, V.E., Knight, P.J., Muench, S.P., Scarff, C.A., White, H.D.(2025) Nature 642: 519-526
- PubMed: 40205053
- DOI: https://doi.org/10.1038/s41586-025-08876-5
- Primary Citation of Related Structures:
8R9V, 8RBF, 8RBG - PubMed Abstract:
Myosins produce force and movement in cells through interactions with F-actin 1 . Generation of movement is thought to arise through actin-catalysed conversion of myosin from an ATP-generated primed (pre-powerstroke) state to a post-powerstroke state, accompanied by myosin lever swing 2,3 . However, the initial, primed actomyosin state has never been observed, and the mechanism by which actin catalyses myosin ATPase activity is unclear. Here, to address these issues, we performed time-resolved cryogenic electron microscopy (cryo-EM) 4 of a myosin-5 mutant having slow hydrolysis product release 5,6 . Primed actomyosin was predominantly captured 10 ms after mixing primed myosin with F-actin, whereas post-powerstroke actomyosin predominated at 120 ms, with no abundant intermediate states detected. For detailed interpretation, cryo-EM maps were fitted with pseudo-atomic models. Small but critical changes accompany the primed motor binding to actin through its lower 50-kDa subdomain, with the actin-binding cleft open and phosphate release prohibited. Amino-terminal actin interactions with myosin promote rotation of the upper 50-kDa subdomain, closing the actin-binding cleft, and enabling phosphate release. The formation of interactions between the upper 50-kDa subdomain and actin creates the strong-binding interface needed for effective force production. The myosin-5 lever swings through 93°, predominantly along the actin axis, with little twisting. The magnitude of lever swing matches the typical step length of myosin-5 along actin 7 . These time-resolved structures demonstrate the swinging lever mechanism, elucidate structural transitions of the power stroke, and resolve decades of conjecture on how myosins generate movement.
- School of Biomedical Sciences, Faculty of Biological Sciences, University of Leeds, Leeds, UK.
Organizational Affiliation: