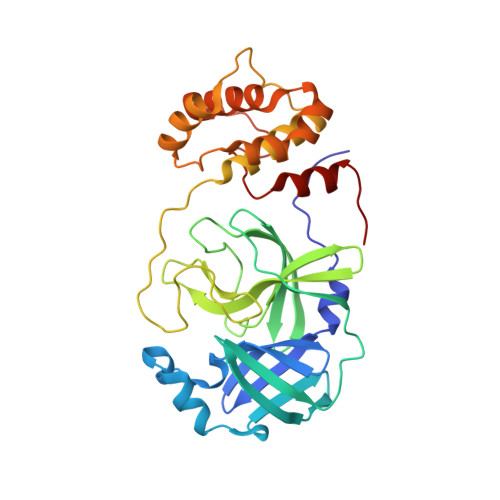
Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Ibrahim, M., Sun, X., Martins de Oliveira, V., Liu, R., Clayton, J., El Kilani, H., Shen, J., Hilgenfeld, R.(2024) Hlife 2: 419-433