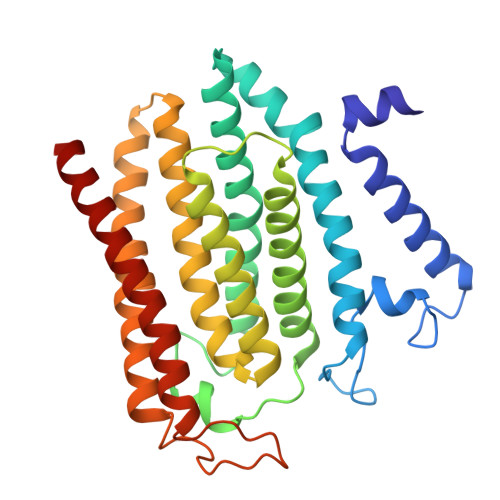
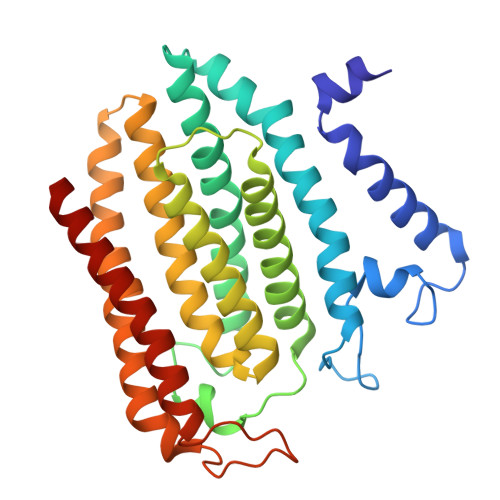
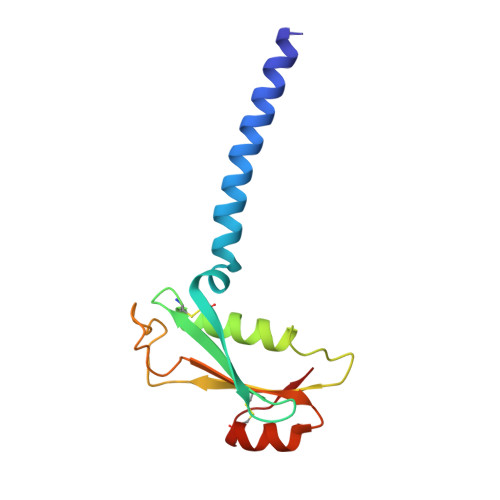
Structure of the yeast ceramide synthase.
Schafer, J.H., Clausmeyer, L., Korner, C., Esch, B.M., Wolf, V.N., Sapia, J., Ahmed, Y., Walter, S., Vanni, S., Januliene, D., Moeller, A., Frohlich, F.(2025) Nat Struct Mol Biol 32: 441-449
- PubMed: 39528796
- DOI: https://doi.org/10.1038/s41594-024-01415-2
- Primary Citation of Related Structures:
8QTN, 8QTR - PubMed Abstract:
Ceramides are essential lipids involved in forming complex sphingolipids and acting as signaling molecules. They result from the N-acylation of a sphingoid base and a CoA-activated fatty acid, a reaction catalyzed by the ceramide synthase (CerS) family of enzymes. Yet, the precise structural details and catalytic mechanisms of CerSs have remained elusive. Here we used cryo-electron microscopy single-particle analysis to unravel the structure of the yeast CerS complex in both an active and a fumonisin B1-inhibited state. Our results reveal the complex's architecture as a dimer of Lip1 subunits bound to the catalytic subunits Lag1 and Lac1. Each catalytic subunit forms a hydrophobic crevice connecting the cytosolic site with the intermembrane space. The active site, located centrally in the tunnel, was resolved in a substrate preloaded state, representing one intermediate in ceramide synthesis. Our data provide evidence for competitive binding of fumonisin B1 to the acyl-CoA-binding tunnel.
- Department of Biology/Chemistry, Structural Biology Section, Osnabrück University, Osnabrück, Germany.
Organizational Affiliation: