Helical aromatic oligoamide foldamers as selective G-quadruplex ligands
Koenig, A., Laffile, V., Largy, E., Thore, S., Mackereth, C., Ferrand, Y., Gabelica, V.(2025) Nucleic Acids Res
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2025) Nucleic Acids Res
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ACE-QUK-QUK-ZY9-QUK | 5 | synthetic construct | Mutation(s): 0 | 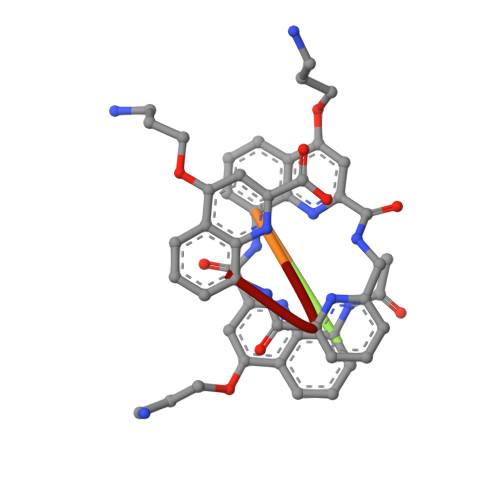 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*TP*GP*GP*GP*TP*TP*GP*GP*GP*TP*TP*GP*GP*GP*TP*TP*GP*GP*GP*T)-3') | 20 | synthetic construct | 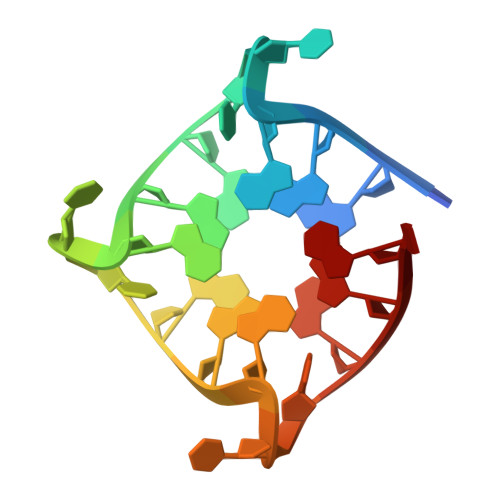 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| K Query on K | D [auth A], E [auth A], F [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | G [auth A], H [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 32.71 | α = 90 |
| b = 32.71 | β = 90 |
| c = 61.01 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Agence Nationale de la Recherche (ANR) | France | ANR-18-CE29-0013 POLYnESI |