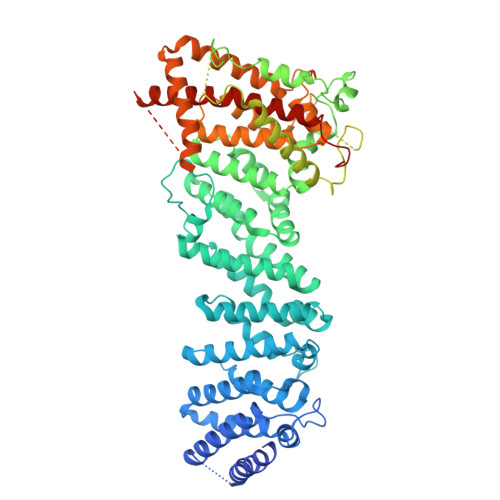
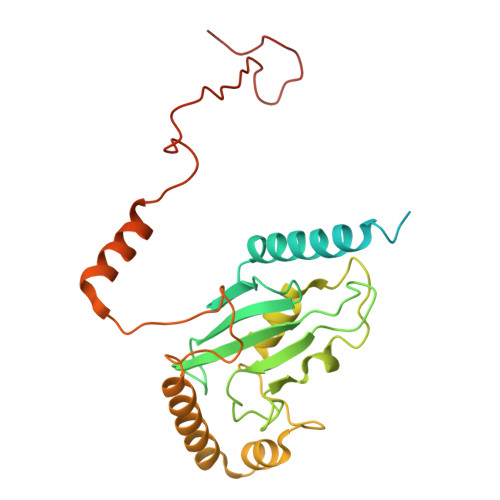
KIF1C activates and extends dynein movement through the FHF cargo adapter.
Abid Ali, F., Zwetsloot, A.J., Stone, C.E., Morgan, T.E., Wademan, R.F., Carter, A.P., Straube, A.(2025) Nat Struct Mol Biol 32: 756-766
- PubMed: 39747486
- DOI: https://doi.org/10.1038/s41594-024-01418-z
- Primary Citation of Related Structures:
8QAT - PubMed Abstract:
Cellular cargos move bidirectionally on microtubules by recruiting opposite polarity motors dynein and kinesin. These motors show codependence, where one requires the activity of the other, although the mechanism is unknown. Here we show that kinesin-3 KIF1C acts as both an activator and a processivity factor for dynein, using in vitro reconstitutions of human proteins. Activation requires only a fragment of the KIF1C nonmotor stalk binding the cargo adapter HOOK3. The interaction site is separate from the constitutive factors FTS and FHIP, which link HOOK3 to small G-proteins on cargos. We provide a structural model for the autoinhibited FTS-HOOK3-FHIP1B (an FHF complex) and explain how KIF1C relieves it. Collectively, we explain codependency by revealing how mutual activation of dynein and kinesin occurs through their shared adapter. Many adapters bind both dynein and kinesins, suggesting this mechanism could be generalized to other bidirectional complexes.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: