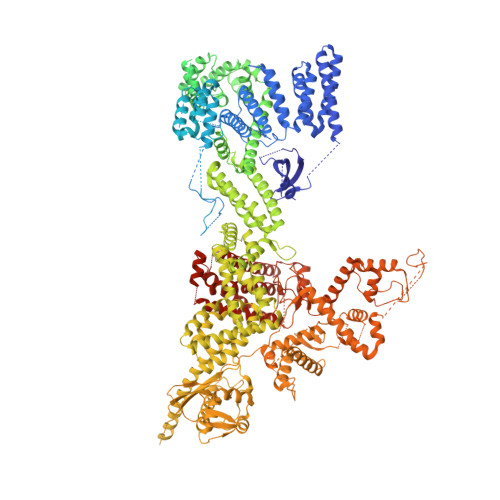
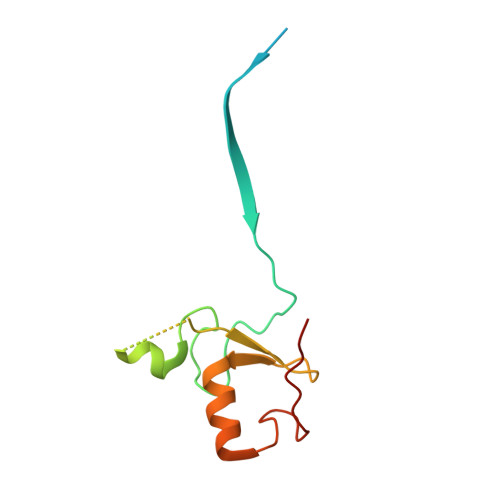
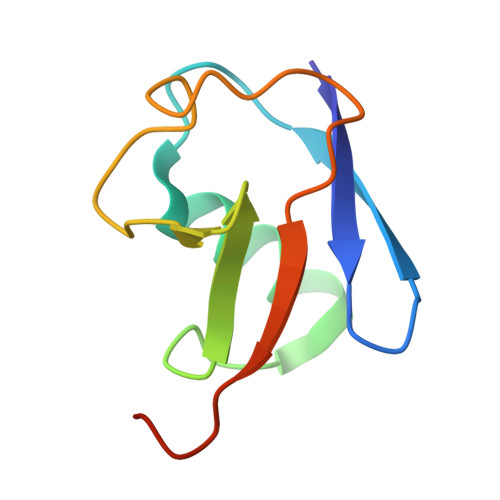
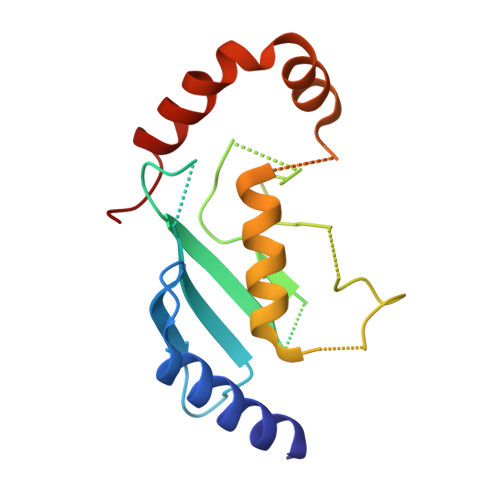
Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Horn-Ghetko, D., Hopf, L.V.M., Tripathi-Giesgen, I., Du, J., Kostrhon, S., Vu, D.T., Beier, V., Steigenberger, B., Prabu, J.R., Stier, L., Bruss, E.M., Mann, M., Xiong, Y., Schulman, B.A.(2024) Nat Struct Mol Biol 31: 1083-1094
- PubMed: 38605244
- DOI: https://doi.org/10.1038/s41594-024-01257-y
- Primary Citation of Related Structures:
8Q7E, 8Q7H, 8RHZ - PubMed Abstract:
Ubiquitin ligation is typically executed by hallmark E3 catalytic domains. Two such domains, 'cullin-RING' and 'RBR', are individually found in several hundred human E3 ligases, and collaborate with E2 enzymes to catalyze ubiquitylation. However, the vertebrate-specific CUL9 complex with RBX1 (also called ROC1), of interest due to its tumor suppressive interaction with TP53, uniquely encompasses both cullin-RING and RBR domains. Here, cryo-EM, biochemistry and cellular assays elucidate a 1.8-MDa hexameric human CUL9-RBX1 assembly. Within one dimeric subcomplex, an E2-bound RBR domain is activated by neddylation of its own cullin domain and positioning from the adjacent CUL9-RBX1 in trans. Our data show CUL9 as unique among RBX1-bound cullins in dependence on the metazoan-specific UBE2F neddylation enzyme, while the RBR domain protects it from deneddylation. Substrates are recruited to various upstream domains, while ubiquitylation relies on both CUL9's neddylated cullin and RBR domains achieving self-assembled and chimeric cullin-RING/RBR E3 ligase activity.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: