Interfacing B-DNA and DNA Mimic Foldamers.
Loos, M., Xu, F., Mandal, P.K., Chakrabortty, T., Douat, C., Konrad, D.B., Cabbar, M., Singer, J., Corvaglia, V., Carell, T., Huc, I.(2025) Angew Chem Int Ed Engl 64: e202505273-e202505273
- PubMed: 40346004
- DOI: https://doi.org/10.1002/anie.202505273
- Primary Citation of Related Structures:
8Q60 - PubMed Abstract:
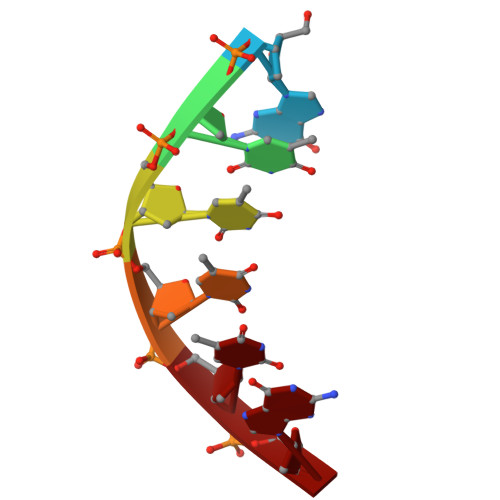
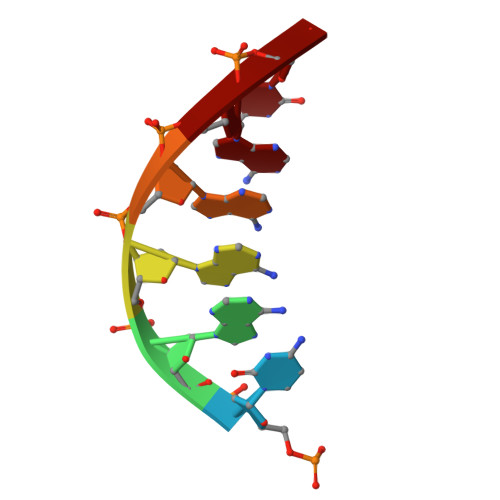
A linker unit was designed and synthesized that can serve both as a hairpin turn in a DNA duplex and anchor point for an aromatic helical foldamer mimicking the shape and surface properties of B-DNA. Methods were developed to synthesize natural/non-natural chimeric molecules combining foldamer and DNA segments. The ability of the linker to position the foldamer helix and the duplex DNA so that their rims and grooves are in register, despite their completely different chemical nature, was demonstrated using single crystal X-ray diffraction, circular dichroism and molecular models. Bio-layer interferometry confirmed that artificial hairpin DNA duplexes keep their ability to bind to DNA binding proteins. The chimeric molecules may pave the way to competitive inhibitors of protein-DNA interactions involving sequence-selective DNA-binding proteins.
- LMU: Ludwig-Maximilians-Universitat Munchen, Pharmacy, GERMANY.
Organizational Affiliation: