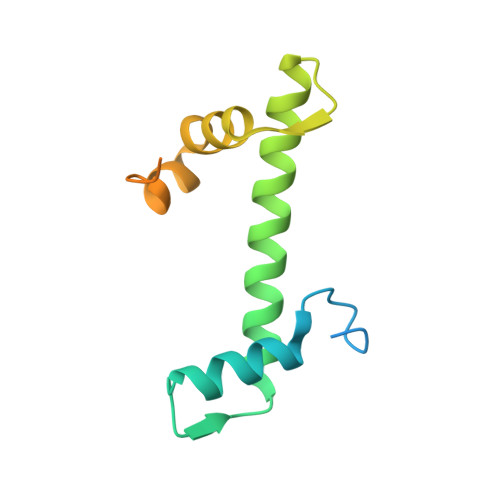
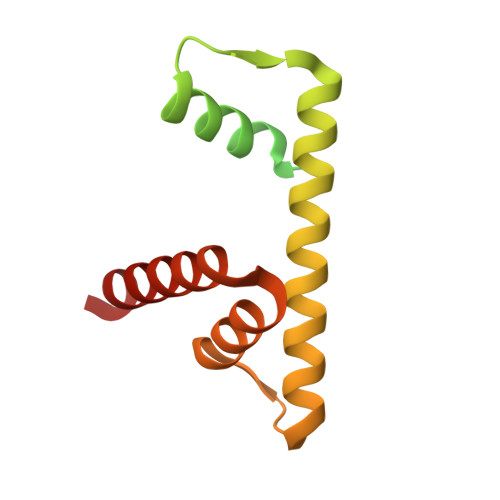
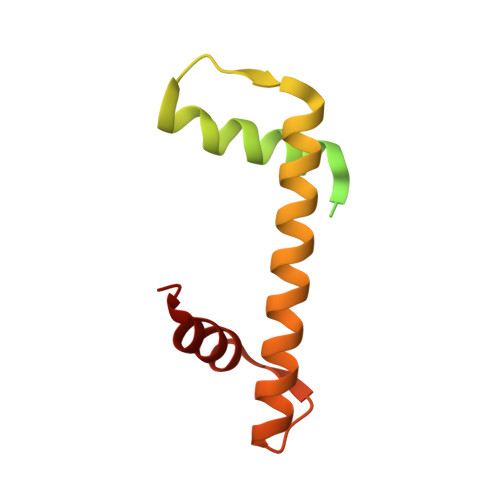
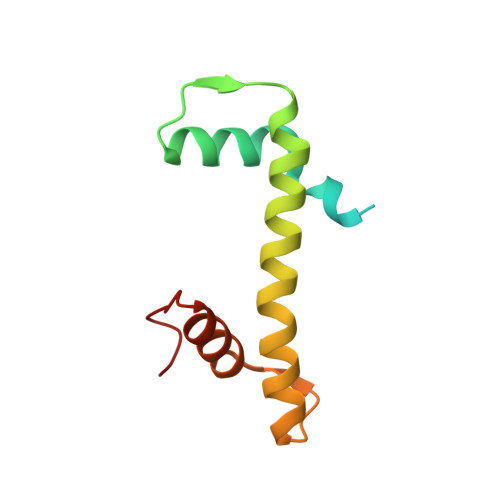
An Oryza-specific histone H4 variant predisposes H4 lysine 5 acetylation to modulate salt stress responses.
Gandhivel, V.H., Sotelo-Parrilla, P., Raju, S., Jha, S., Gireesh, A., Harshith, C.Y., Gut, F., Vinothkumar, K.R., Berger, F., Jeyaprakash, A.A., Shivaprasad, P.V.(2025) Nat Plants 11: 790-807
- PubMed: 40200022
- DOI: https://doi.org/10.1038/s41477-025-01974-2
- Primary Citation of Related Structures:
8Q15, 8Q16 - PubMed Abstract:
Paralogous variants of canonical histones guide accessibility to DNA and function as additional layers of genome regulation. Across eukaryotes, the mechanism of action and functional significance of several variants of core histones are well known except those of histone H4. Here we show that a variant of H4 (H4.V) expressing tissue-specifically among Oryza members mediated specific epigenetic changes contributing to salt tolerance. H4.V was incorporated into specific heterochromatic sites, where it blocked the deposition of active histone marks. Stress-dependent redistribution of H4.V enabled the incorporation of acetylated H4 lysine 5 (H4K5ac) in the gene bodies. The misexpression of H4.V led to defects in reproductive development and in mounting salt stress responses. H4.V formed homotypic nucleosomes and mediated these alterations by conferring distinct molecular properties to the nucleosomes, as seen with cryo electron microscopy structures and biochemical assays. These results reveal not only an H4 variant among plants but also a chromatin regulation that might have contributed to the adaptation of semi-aquatic Oryza members.
- National Centre for Biological Sciences, TIFR, GKVK Campus, Bangalore, India.
Organizational Affiliation: