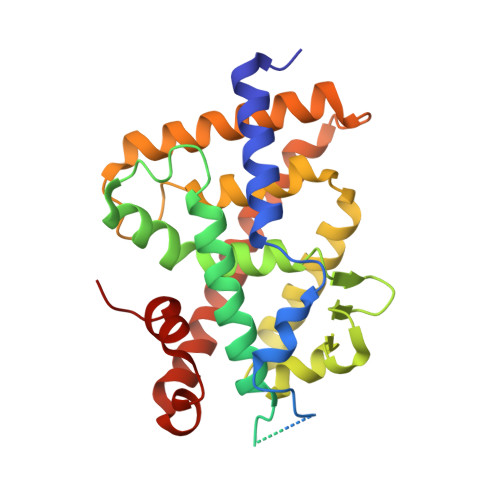
Further Studies on the Highly Active Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3 (Calcitriol): Refinement of the Side Chain.
Zarate-Ruiz, A., Seoane, S., Peluso-Iltis, C., Peters, S., Gregorio, C., Guiberteau, T., Maestro, M., Perez-Fernandez, R., Rochel, N., Mourino, A.(2023) J Med Chem 66: 15326-15339
- PubMed: 37910811
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01371
- Primary Citation of Related Structures:
8PWD, 8PWE, 8PWF, 8PWM - PubMed Abstract:
Current efforts in the vitamin D field are directed toward the development of highly antiproliferative yet noncalcemic analogues of the natural hormone 1α,25-dihydroxyvitamin D 3 (1,25D 3 ). We have recently reported the design, synthesis, biological evaluation, and crystal structures of a series of novel analogues that both lack the steroidal C-ring and have an m -phenylene ring replacing the steroidal cyclopentane D-ring. We have now investigated the potentiating effects of incorporating selected modifications (hexafluorination and/or an internal triple bond) within the steroidal side chain in our series. An alternative synthetic strategy (Wittig-Horner approach instead of our previously used Pd-catalyzed tandem cyclization/cross-coupling) for the construction of the vitamin D triene system was found convenient for the target compounds 2 , 3a , 3b , and 3c of this report. These modifications enhance vitamin D nuclear receptor (VDR) interactions and consequently VDR-associated biological properties compared to parental PG-136 compound while maintaining normal calcium levels.
- Department of Organic Chemistry, Research Laboratory Ignacio Ribas, University of Santiago de Compostela, Avda. de las Ciencias s/n, Santiago de Compostela 15782, Spain.
Organizational Affiliation: