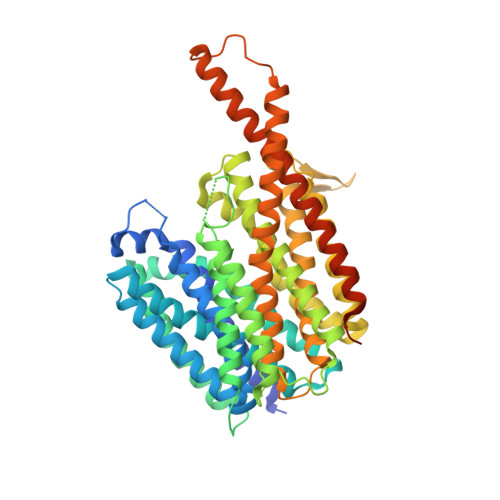
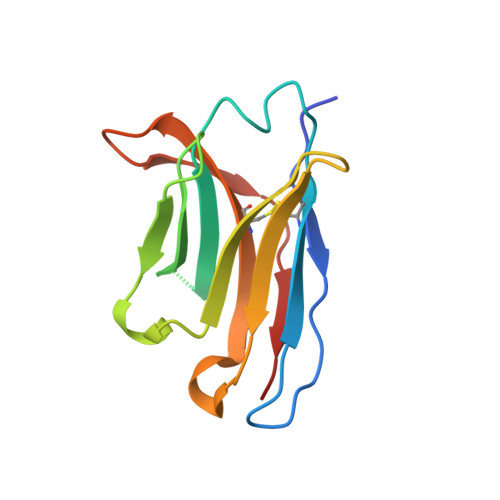
Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410.
Remm, S., De Vecchis, D., Schoppe, J., Hutter, C.A.J., Gonda, I., Hohl, M., Newstead, S., Schafer, L.V., Seeger, M.A.(2023) Nat Commun 14: 6449-6449
- PubMed: 37833269
- DOI: https://doi.org/10.1038/s41467-023-42073-0
- Primary Citation of Related Structures:
8PNL - PubMed Abstract:
Mycobacterium tuberculosis is protected from antibiotic therapy by a multi-layered hydrophobic cell envelope. Major facilitator superfamily (MFS) transporter Rv1410 and the periplasmic lipoprotein LprG are involved in transport of triacylglycerides (TAGs) that seal the mycomembrane. Here, we report a 2.7 Å structure of a mycobacterial Rv1410 homologue, which adopts an outward-facing conformation and exhibits unusual transmembrane helix 11 and 12 extensions that protrude ~20 Å into the periplasm. A small, very hydrophobic cavity suitable for lipid transport is constricted by a functionally important ion-lock likely involved in proton coupling. Combining mutational analyses and MD simulations, we propose that TAGs are extracted from the core of the inner membrane into the central cavity via lateral clefts present in the inward-facing conformation. The functional role of the periplasmic helix extensions is to channel the extracted TAG into the lipid binding pocket of LprG.
- Institute of Medical Microbiology, University of Zurich, Zürich, Switzerland.
Organizational Affiliation: