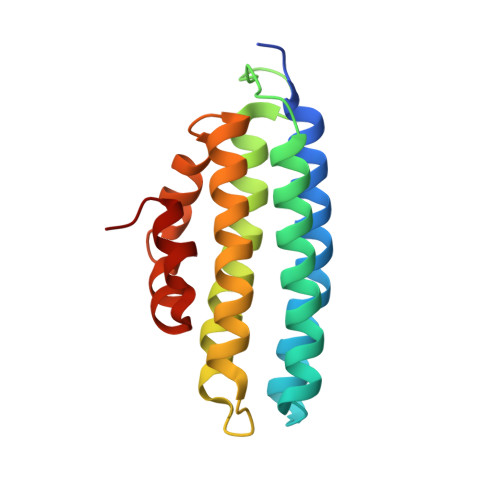
Role and structure of the small subunit forming heterodimers with laccase-like enzymes.
Aza, P., Linde, D., Molpeceres, G., Vind, J., Medrano, F.J., Camarero, S.(2023) Protein Sci 32: e4734-e4734
- PubMed: 37483125
- DOI: https://doi.org/10.1002/pro.4734
- Primary Citation of Related Structures:
8PAQ - PubMed Abstract:
Unlike laccases sensu stricto, which are usually monomeric enzymes, laccase-like enzymes recently re-classified as Novel Laccases (NLACs) are characterized by the formation of heterodimers with small proteins (subunits) of unknown function. Here the NLAC from Pleurotus eryngii (PeNL) and a small protein selected from the fungal genome, that is homologous to reported POXA3 from Pleurotus ostreatus, were produced in Aspergillus oryzae separately or together. The two proteins interacted regardless of whether the small subunit was co-expressed or exogenously added to the enzyme. The stability and catalytic activity of PeNL was significantly enhanced in the presence of the small subunit. Size exclusion chromatography-multi angle light scattering (SEC-MALS) analysis confirmed that the complex PeNL-ss is a heterodimer of 77.4 kDa. The crystallographic structure of the small protein expressed in Escherichia coli was solved at 1.6 Å resolution. This is the first structure elucidated of a small subunit of a NLAC. The helix bundle structure of the small subunit accommodates well with the enzyme model structure, including interactions with specific regions of NLACs and some amino acid residues of the substrate-binding loops.
- Centro de Investigaciones Biológicas Margarita Salas, CSIC, Madrid, Spain.
Organizational Affiliation: