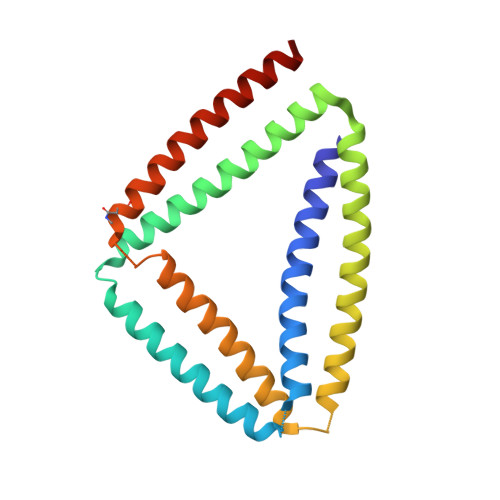
Crystal Structure of de Novo Designed Coiled-Coil Protein Origami Triangle.
Satler, T., Hadzi, S., Jerala, R.(2023) J Am Chem Soc 145: 16995-17000
- PubMed: 37486611
- DOI: https://doi.org/10.1021/jacs.3c05531
- Primary Citation of Related Structures:
8P4Y - PubMed Abstract:
Coiled-coil protein origami (CCPO) uses modular coiled-coil building blocks and topological principles to design polyhedral structures distinct from those of natural globular proteins. While the CCPO strategy has proven successful in designing diverse protein topologies, no high-resolution structural information has been available about these novel protein folds. Here we report the crystal structure of a single-chain CCPO in the shape of a triangle. While neither cyclization nor the addition of nanobodies enabled crystallization, it was ultimately facilitated by the inclusion of a GCN 2 homodimer. Triangle edges are formed by the orthogonal parallel coiled-coil dimers P1:P2, P3:P4, and GCN 2 connected by short linkers. A triangle has a large central cavity and is additionally stabilized by side-chain interactions between neighboring segments at each vertex. The crystal lattice is densely packed and stabilized by a large number of contacts between triangles. Interestingly, the polypeptide chain folds into a trefoil-type protein knot topology, and AlphaFold2 fails to predict the correct fold. The structure validates the modular CC-based protein design strategy, providing molecular insight underlying CCPO stabilization and new opportunities for the design.
- Department of Synthetic Biology and Immunology, National Institute of Chemistry, 1000 Ljubljana, Slovenia.
Organizational Affiliation: