Thogoto virus polymerase
Kouba, T., Cusack, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polymerase acidic protein | 622 | Thogotovirus thogotoense | Mutation(s): 0 Gene Names: Segment 3 | 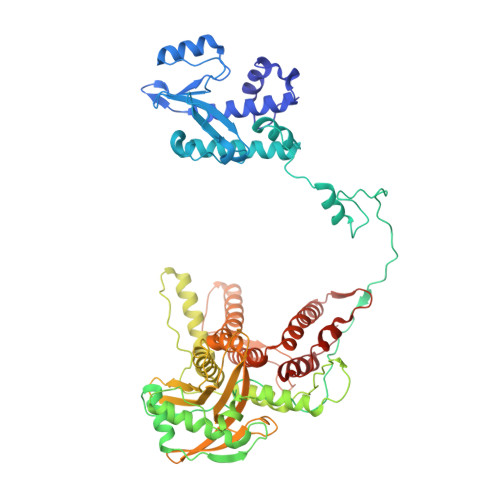 | |
UniProt | |||||
Find proteins for P27194 (Thogoto virus (isolate SiAr 126)) Explore P27194 Go to UniProtKB: P27194 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27194 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-directed RNA polymerase catalytic subunit | 710 | Thogotovirus thogotoense | Mutation(s): 0 Gene Names: Segment 2 EC: 2.7.7.48 | 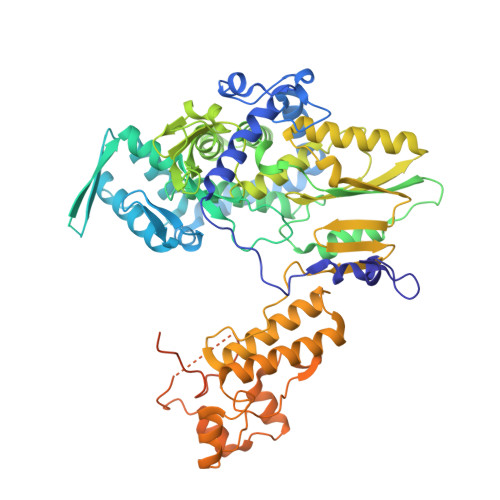 | |
UniProt | |||||
Find proteins for O41353 (Thogoto virus (isolate SiAr 126)) Explore O41353 Go to UniProtKB: O41353 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O41353 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polymerase basic protein 2 | 769 | Thogotovirus thogotoense | Mutation(s): 0 Gene Names: Segment 1 | 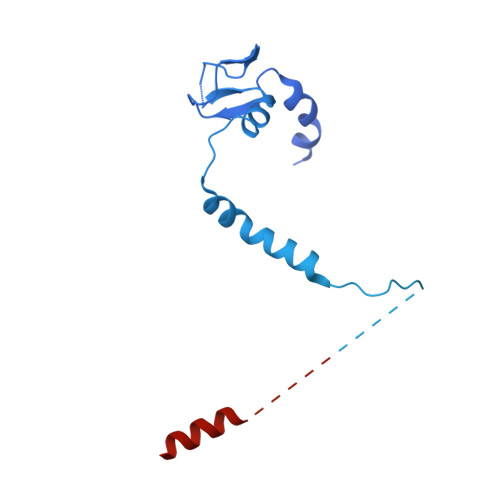 | |
UniProt | |||||
Find proteins for Q9YNA4 (Thogoto virus (isolate SiAr 126)) Explore Q9YNA4 Go to UniProtKB: Q9YNA4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9YNA4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 3'RNA | D [auth R], E [auth V] | 32 | Thogotovirus thogotoense | 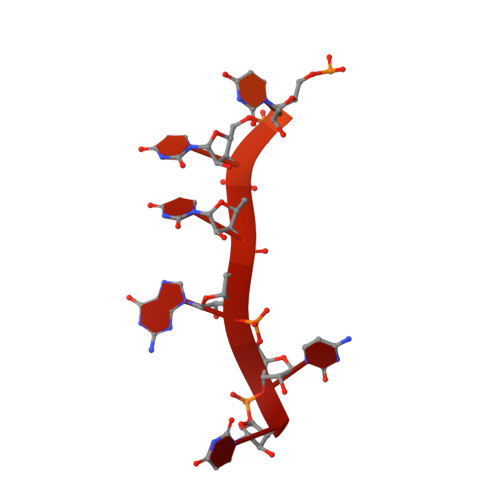 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | F [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| French National Research Agency | France | -- |