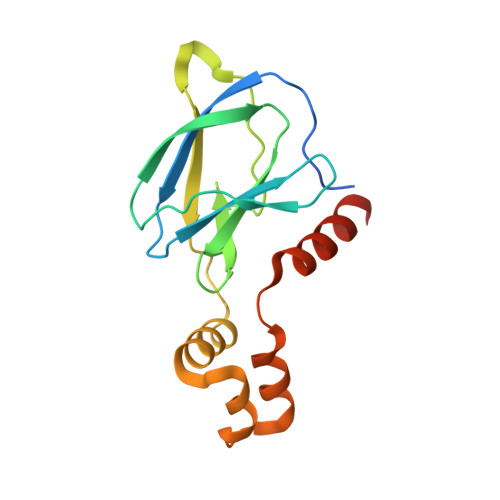
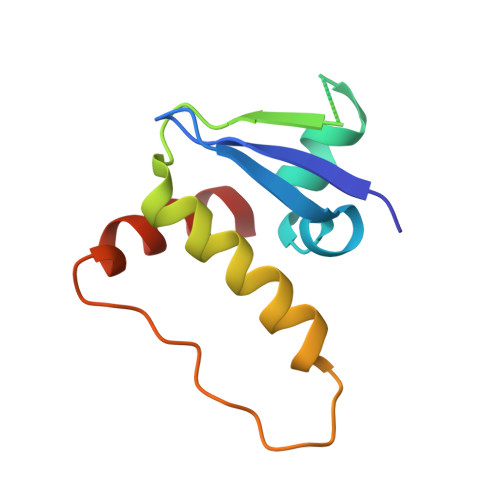
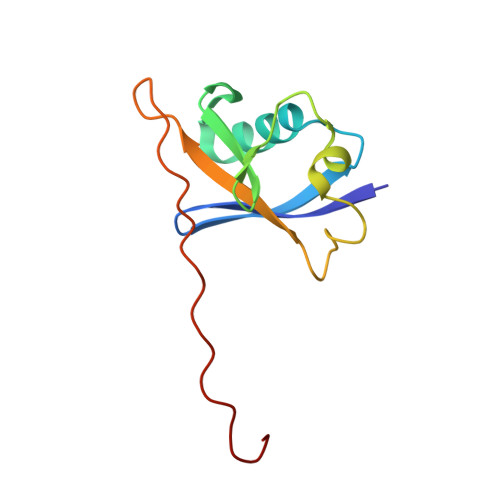
Drugit: crowd-sourcing molecular design of non-peptidic VHL binders.
Scott, T., Smethurst, C.A.P., Westermaier, Y., Mayer, M., Greb, P., Kousek, R., Biberger, T., Bader, G., Jandova, Z., Schmalhorst, P.S., Fuchs, J.E., Magarkar, A., Hoenke, C., Gerstberger, T., Combs, S.A., Pape, R., Phul, S., Kothiwale, S., Bergner, A., Waterson, A.G., Weinstabl, H., McConnell, D.B., Meiler, J., Bottcher, J., Moretti, R.(2025) Nat Commun 16: 3548-3548
- PubMed: 40229246
- DOI: https://doi.org/10.1038/s41467-025-58406-0
- Primary Citation of Related Structures:
8P0F - PubMed Abstract:
Building on the role of human intuition in small molecule drug design, we explored whether crowdsourcing could recruit citizen scientists to this task while in parallel building awareness for this scientific process. Here, we introduce Drugit ( https://drugit.org ), the small molecule design mode of the online citizen science game Foldit. We demonstrate its utility by identifying distinct binders to the von Hippel Lindau E3 ligase. Several thousand molecules were suggested by players in a series of ten puzzle rounds. The proposed molecules were further evaluated in silico and manually by an expert panel. Selected candidates were synthesized and tested. One of these molecules shows dose-dependent shift perturbations in protein-observed NMR experiments. The co-crystal structure in complex with the E3 ligase reveals that the observed binding mode matches the player's original idea. The completion of one full design cycle is a proof of concept for the Drugit approach and highlights the potential of involving citizen scientists in early drug discovery.
- Department of Chemistry, Vanderbilt University, Nashville, TN, 37235, USA.
Organizational Affiliation: