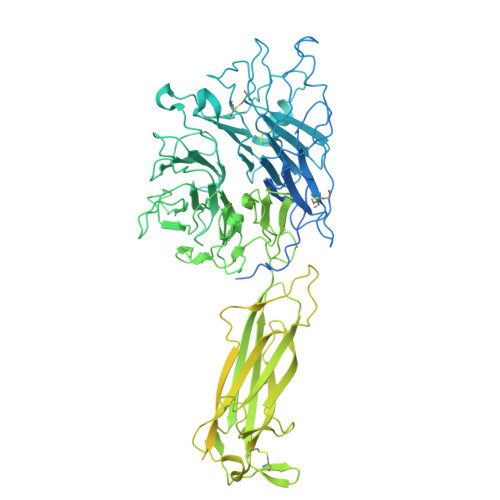
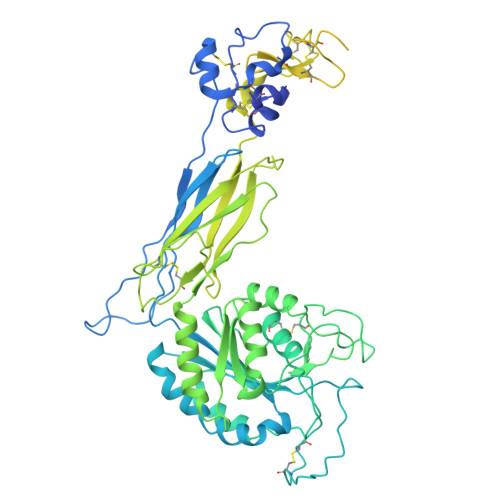
Spatial N-glycan rearrangement on alpha 5 beta 1 integrin nucleates galectin-3 oligomers to determine endocytic fate.
Shafaq-Zadah, M., Dransart, E., Hamitouche, I., Wunder, C., Chambon, V., Valades-Cruz, C.A., Leconte, L., Sarangi, N.K., Robinson, J., Bai, S.K., Regmi, R., Di Cicco, A., Hovasse, A., Bartels, R., Nilsson, U.J., Cianferani-Sanglier, S., Leffler, H., Keyes, T.E., Levy, D., Raunser, S., Roderer, D., Johannes, L.(2025) Nat Commun 16: 9461-9461
- PubMed: 41145507
- DOI: https://doi.org/10.1038/s41467-025-64523-7
- Primary Citation of Related Structures:
8OXZ - PubMed Abstract:
Membrane glycoproteins frequently adopt different conformations when altering between active and inactive states. Here, we discover a molecular switch that exploits dynamic spatial rearrangements of N-glycans during such conformational transitions to control protein function. For the conformationally switchable cell adhesion glycoprotein α 5 β 1 integrin, we find that only the bent-closed state arranges N-glycans to nucleate the formation of up to tetrameric oligomers of the glycan-binding protein galectin-3. We propose a structural model of how these galectin-3 oligomers are built and how they clamp the bent-closed state to select it for endocytic uptake and subsequent retrograde trafficking to the Golgi for polarized distribution in cells. Our findings reveal the dynamic regulation of the glycan landscape at the cell surface to achieve oligomerization of galectin-3. Galectin-3 oligomers are thereby identified as functional decoders of defined spatial patterns of N-glycans on specifically the bent-closed conformational state of α 5 β 1 integrin and possibly other integrin family members.
- Chemical Biology of Cancer Unit, Institut Curie, U1339 INSERM, UMR3666 CNRS, PSL Research University, Paris, France. massiullah.shafaq-zadah@curie.fr.
Organizational Affiliation: