Targeted protein degradation via intramolecular bivalent glues.
Hsia, O., Hinterndorfer, M., Cowan, A.D., Iso, K., Ishida, T., Sundaramoorthy, R., Nakasone, M.A., Imrichova, H., Schatz, C., Rukavina, A., Husnjak, K., Wegner, M., Correa-Saez, A., Craigon, C., Casement, R., Maniaci, C., Testa, A., Kaulich, M., Dikic, I., Winter, G.E., Ciulli, A.(2024) Nature 627: 204-211
- PubMed: 38383787
- DOI: https://doi.org/10.1038/s41586-024-07089-6
- Primary Citation of Related Structures:
8OV6 - PubMed Abstract:
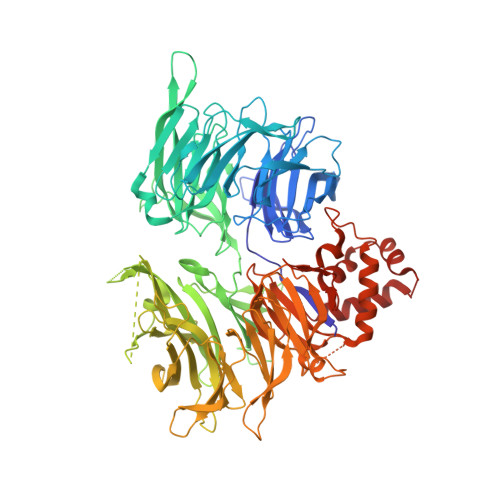
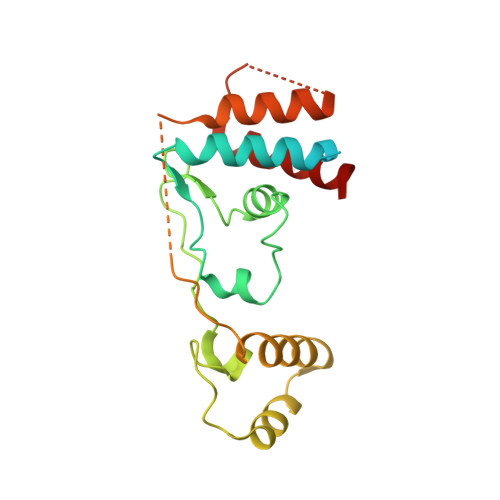
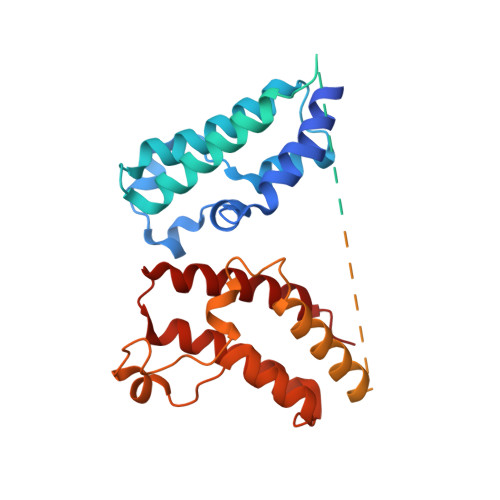
Targeted protein degradation is a pharmacological modality that is based on the induced proximity of an E3 ubiquitin ligase and a target protein to promote target ubiquitination and proteasomal degradation. This has been achieved either via proteolysis-targeting chimeras (PROTACs)-bifunctional compounds composed of two separate moieties that individually bind the target and E3 ligase, or via molecular glues that monovalently bind either the ligase or the target 1-4 . Here, using orthogonal genetic screening, biophysical characterization and structural reconstitution, we investigate the mechanism of action of bifunctional degraders of BRD2 and BRD4, termed intramolecular bivalent glues (IBGs), and find that instead of connecting target and ligase in trans as PROTACs do, they simultaneously engage and connect two adjacent domains of the target protein in cis. This conformational change 'glues' BRD4 to the E3 ligases DCAF11 or DCAF16, leveraging intrinsic target-ligase affinities that do not translate to BRD4 degradation in the absence of compound. Structural insights into the ternary BRD4-IBG1-DCAF16 complex guided the rational design of improved degraders of low picomolar potency. We thus introduce a new modality in targeted protein degradation, which works by bridging protein domains in cis to enhance surface complementarity with E3 ligases for productive ubiquitination and degradation.
- Centre for Targeted Protein Degradation, School of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: