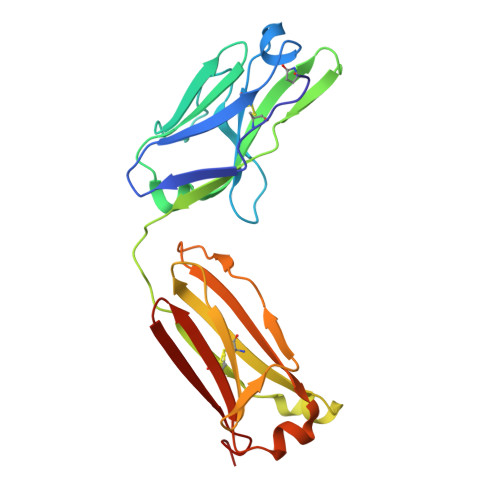
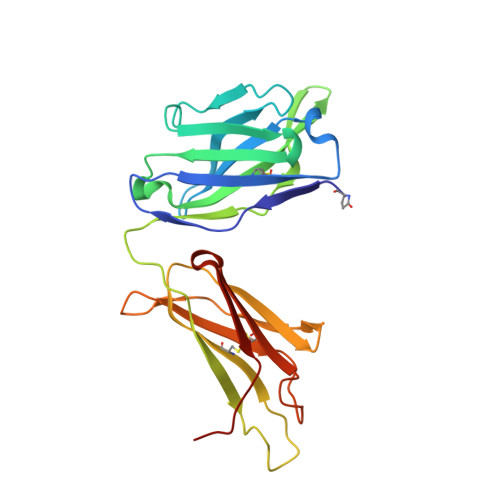
Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Davies, A.M., Beavil, R.L., Barbolov, M., Sandhar, B.S., Gould, H.J., Beavil, A.J., Sutton, B.J., McDonnell, J.M.(2023) Mol Immunol 159: 28-37
- PubMed: 37267832
- DOI: https://doi.org/10.1016/j.molimm.2023.05.006
- Primary Citation of Related Structures:
8OJS, 8OJT, 8OJU, 8OJV - PubMed Abstract:
Antibodies of the IgD isotype remain the least well characterized of the mammalian immunoglobulin isotypes. Here we report three-dimensional structures for the Fab region of IgD, based on four different crystal structures, at resolutions of 1.45-2.75 Å. These IgD Fab crystals provide the first high-resolution views of the unique Cδ1 domain. Structural comparisons identify regions of conformational diversity within the Cδ1 domain, as well as among the homologous domains of Cα1, Cγ1 and Cμ1. The IgD Fab structure also possesses a unique conformation of the upper hinge region, which may contribute to the overall disposition of the very long linker sequence between the Fab and Fc regions found in human IgD. Structural similarities observed between IgD and IgG, and differences with IgA and IgM, are consistent with predicted evolutionary relationships for the mammalian antibody isotypes.
- King's College London, Randall Centre for Cell and Molecular Biophysics, New Hunt's House, London SE1 1UL, United Kingdom.
Organizational Affiliation: