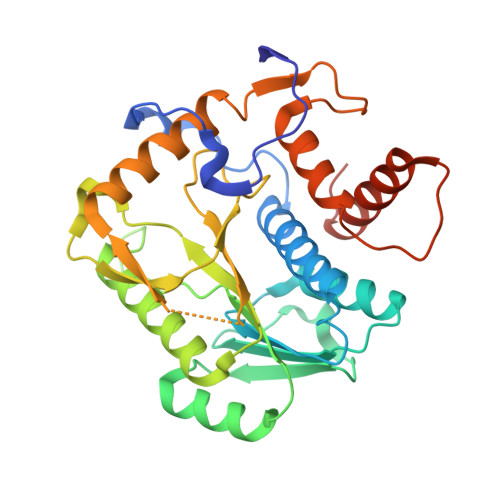
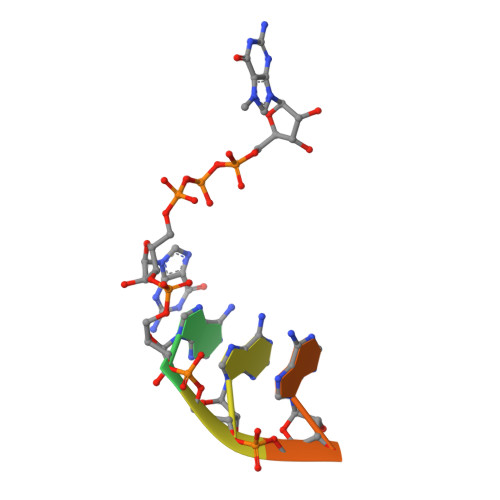
Structural basis for RNA-cap recognition and methylation by the mpox methyltransferase VP39.
Skvara, P., Chalupska, D., Klima, M., Kozic, J., Silhan, J., Boura, E.(2023) Antiviral Res 216: 105663-105663
- PubMed: 37421984
- DOI: https://doi.org/10.1016/j.antiviral.2023.105663
- Primary Citation of Related Structures:
8OIV - PubMed Abstract:
Mpox is a zoonotic disease caused by the mpox virus (MPXV), which has gained attention due to its rapid and widespread transmission, with reports from more than 100 countries. The virus belongs to the Orthopoxvirus genus, which also includes variola virus and vaccinia virus. In poxviruses, the RNA cap is crucial for the translation and stability of viral mRNAs and also for immune evasion. This study presents the crystal structure of the mpox 2'-O-methyltransfarase VP39 in complex with a short cap-0 RNA. The RNA substrate binds to the protein without causing any significant changes to its overall fold and is held in place by a combination of electrostatic interactions, π-π stacking and hydrogen bonding. The structure also explains the mpox VP39 preference for a guanine base at the first position; it reveals that guanine forms a hydrogen bond that an adenine would not be able to form.
- Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, V.v.i, Flemingovo nám. 2, 16610, Prague 6, Czech Republic.
Organizational Affiliation: