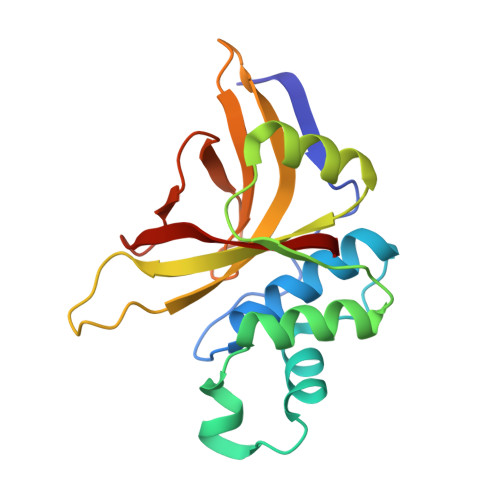
Crystal Structure of Staphopain C from Staphylococcus aureus.
Magoch, M., McEwen, A.G., Napolitano, V., Wladyka, B., Dubin, G.(2023) Molecules 28
- PubMed: 37298883
- DOI: https://doi.org/10.3390/molecules28114407
- Primary Citation of Related Structures:
8OIG - PubMed Abstract:
Staphylococcus aureus is a common opportunistic pathogen of humans and livestock that causes a wide variety of infections. The success of S. aureus as a pathogen depends on the production of an array of virulence factors including cysteine proteases (staphopains)-major secreted proteases of certain strains of the bacterium. Here, we report the three-dimensional structure of staphopain C (ScpA2) of S. aureus , which shows the typical papain-like fold and uncovers a detailed molecular description of the active site. Because the protein is involved in the pathogenesis of a chicken disease, our work provides the foundation for inhibitor design and potential antimicrobial strategies against this pathogen.
- Malopolska Centre of Biotechnology, Jagiellonian University, 30-387 Krakow, Poland.
Organizational Affiliation: