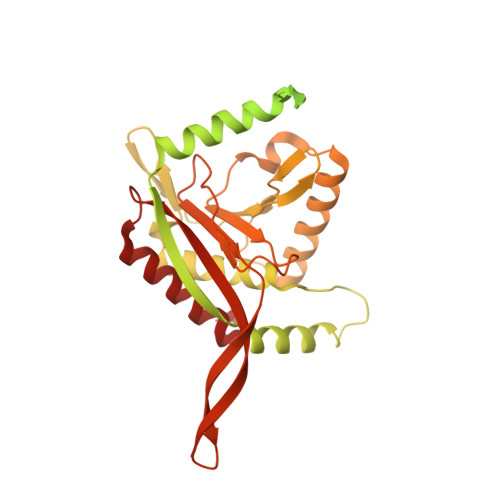
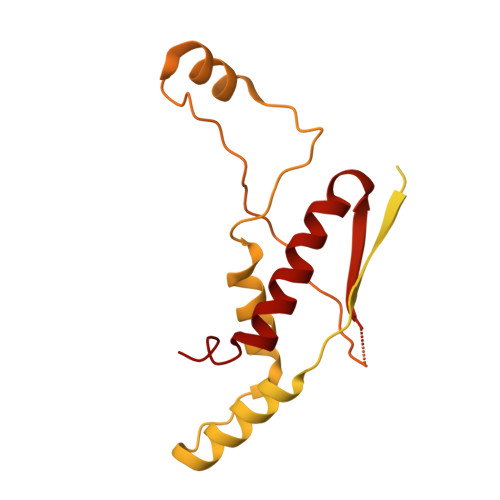
The structure and evolutionary diversity of the fungal E3-binding protein.
Forsberg, B.O.(2023) Commun Biol 6: 480-480
- PubMed: 37137945
- DOI: https://doi.org/10.1038/s42003-023-04854-7
- Primary Citation of Related Structures:
7R5M, 8OHS - PubMed Abstract:
The pyruvate dehydrogenase complex (PDC) is a central metabolic enzyme in all living cells composed majorly of E1, E2, and E3. Tight coupling of their reactions makes each component essential, so that any loss impacts oxidative metabolism pathologically. E3 retention is mediated by the E3-binding protein (E3BP), which is here resolved within the PDC core from N.crassa, resolved to 3.2Å. Fungal and mammalian E3BP are shown to be orthologs, arguing E3BP as a broadly eukaryotic gene. Fungal E3BP architectures predicted from sequence data and computational models further bridge the evolutionary distance between N.crassa and humans, and suggest discriminants for E3-specificity. This is confirmed by similarities in their respective E3-binding domains, where an interaction previously not described is also predicted. This provides evolutionary parallels for a crucial interaction human metabolism, an interaction specific to fungi that can be targeted, and an example of protein evolution following gene neofunctionalization.
- Department of Physiology and Pharmacology, Karolinska Institutet, Biomedicum, Solnavägen 9, 171 77, Stockholm, Sweden. bjorn.forsberg@ki.se.
Organizational Affiliation: