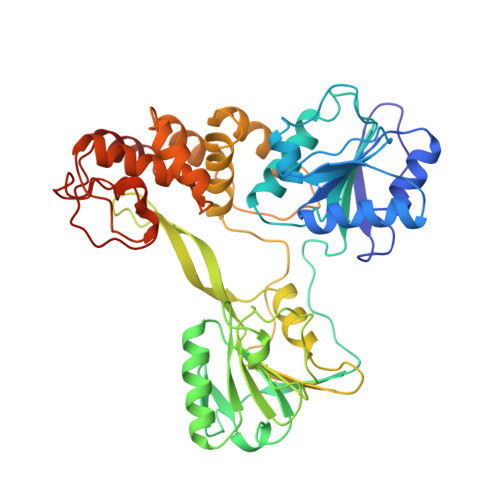
Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA.
Cho, H.S., Ha, N.C., Kang, L.W., Chung, K.M., Back, S.H., Jang, S.K., Oh, B.H.(1998) J Biological Chem 273: 15045-15052
- PubMed: 9614113
- DOI: https://doi.org/10.1074/jbc.273.24.15045
- Primary Citation of Related Structures:
8OHM - PubMed Abstract:
Crystal structure of RNA helicase domain from genotype 1b hepatitis C virus has been determined at 2.3 A resolution by the multiple isomorphous replacement method. The structure consists of three domains that form a Y-shaped molecule. One is a NTPase domain containing two highly conserved NTP binding motifs. Another is an RNA binding domain containing a conserved RNA binding motif. The third is a helical domain that contains no beta-strand. The RNA binding domain of the molecule is distinctively separated from the other two domains forming an interdomain cleft into which single stranded RNA can be modeled. A channel is found between a pair of symmetry-related molecules which exhibit the most extensive crystal packing interactions. A stretch of single stranded RNA can be modeled with electrostatic complementarity into the interdomain cleft and continuously through the channel. These observations suggest that some form of this dimer is likely to be the functional form that unwinds double stranded RNA processively by passing one strand of RNA through the channel and passing the other strand outside of the dimer. A "descending molecular see-saw" model is proposed that is consistent with directionality of unwinding and other physicochemical properties of RNA helicases.
- Department of Life Sciences, Pohang University of Science and Technology, Pohang, Kyungbuk, 790-784, South Korea.
Organizational Affiliation: