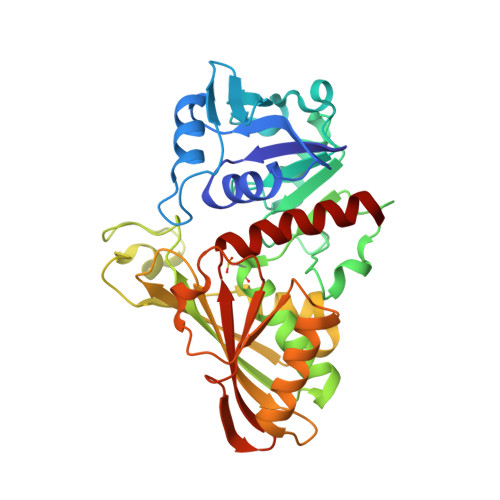
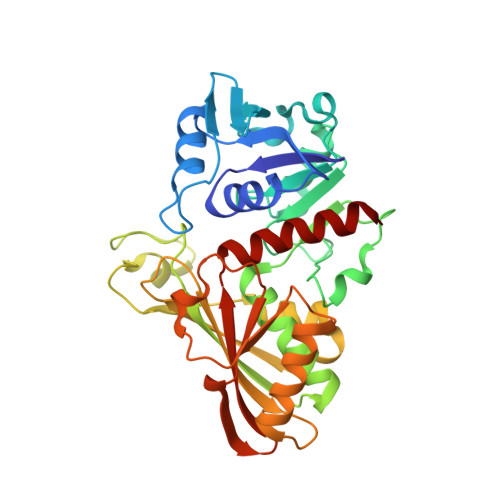
The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Navas-Yuste, S., de la Paz, K., Querol-Garcia, J., Gomez-Quevedo, S., Rodriguez de Cordoba, S., Fernandez, F.J., Vega, M.C.(2023) Front Immunol 14: 1190943-1190943
- PubMed: 37409124
- DOI: https://doi.org/10.3389/fimmu.2023.1190943
- Primary Citation of Related Structures:
8OHA - PubMed Abstract:
Leptospirosis is a neglected worldwide zoonosis involving farm animals and domestic pets caused by the Gram-negative spirochete Leptospira interrogans . This bacterium deploys a variety of immune evasive mechanisms, some of them targeted at the complement system of the host's innate immunity. In this work, we have solved the X-ray crystallographic structure of L. interrogans glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to 2.37-Å resolution, a glycolytic enzyme that has been shown to exhibit moonlighting functions that potentiate infectivity and immune evasion in various pathogenic organisms. Besides, we have characterized the enzyme's kinetic parameters toward the cognate substrates and have proven that the two natural products anacardic acid and curcumin are able to inhibit L. interrogans GAPDH at micromolar concentration through a noncompetitive inhibition modality. Furthermore, we have established that L. interrogans GAPDH can interact with the anaphylatoxin C5a of human innate immunity in vitro using bio-layer interferometry and a short-range cross-linking reagent that tethers free thiol groups in protein complexes. To shed light into the interaction between L. interrogans GAPDH and C5a, we have also carried out cross-link guided protein-protein docking. These results suggest that L. interrogans could be placed in the growing list of bacterial pathogens that exploit glycolytic enzymes as extracellular immune evasive factors. Analysis of the docking results indicates a low affinity interaction that is consistent with previous evidence, including known binding modes of other α-helical proteins with GAPDH. These findings allow us to propose L. interrogans GAPDH as a potential immune evasive factor targeting the complement system.
- Centro de Investigaciones Biológicas Margarita Salas, Consejo Superior de Investigaciones Científicas (CSIC), Madrid, Spain.
Organizational Affiliation: