Structural insights into the human HRD1 ubiquitin ligase complex
Guo, L., Liu, G., He, J., Jia, X., He, Y., Wang, Z., Qian, H.(2025) Nat Commun 16: 6007
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein sel-1 homolog 1 | A [auth D], D [auth C] | 794 | Homo sapiens | Mutation(s): 0 Gene Names: SEL1L, TSA305, UNQ128/PRO1063 Membrane Entity: Yes | 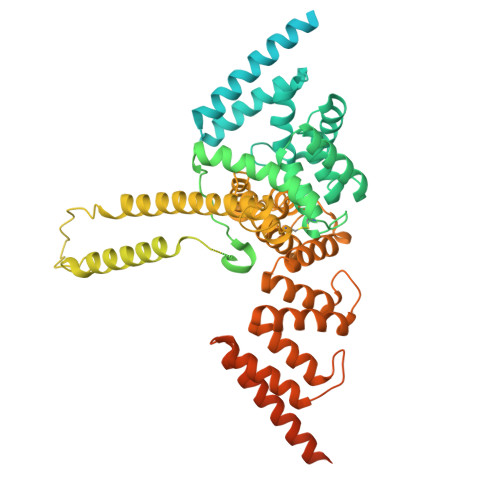 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UBV2 (Homo sapiens) Explore Q9UBV2 Go to UniProtKB: Q9UBV2 | |||||
PHAROS: Q9UBV2 GTEx: ENSG00000071537 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UBV2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | Go to GlyGen: Q9UBV2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ala-Asn-Ala | B [auth G], E [auth H] | 3 | Homo sapiens | Mutation(s): 0 | 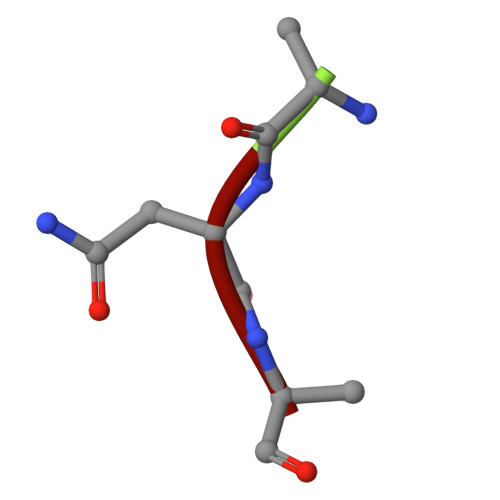 |
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endoplasmic reticulum lectin 1 | C [auth E], F | 483 | Homo sapiens | Mutation(s): 0 Gene Names: ERLEC1, C2orf30, XTP3TPB, UNQ1878/PRO4321 Membrane Entity: Yes | 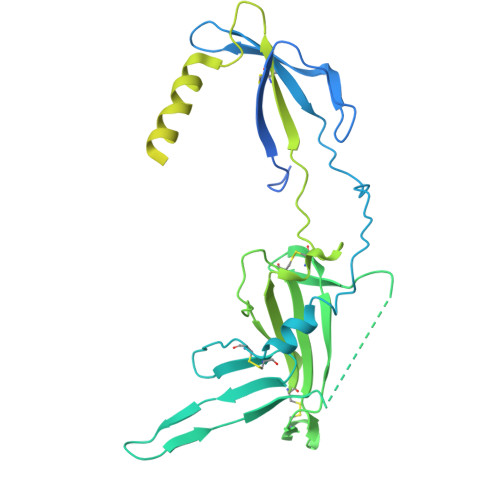 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96DZ1 (Homo sapiens) Explore Q96DZ1 Go to UniProtKB: Q96DZ1 | |||||
PHAROS: Q96DZ1 GTEx: ENSG00000068912 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96DZ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| E3 ubiquitin-protein ligase synoviolin | G [auth A], H [auth B] | 617 | Homo sapiens | Mutation(s): 0 Gene Names: SYVN1, HRD1, KIAA1810 EC: 2.3.2.27 Membrane Entity: Yes | 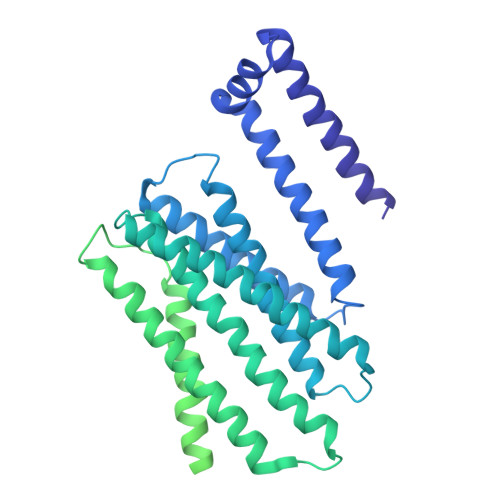 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q86TM6 (Homo sapiens) Explore Q86TM6 Go to UniProtKB: Q86TM6 | |||||
PHAROS: Q86TM6 GTEx: ENSG00000162298 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86TM6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | K [auth D] L [auth D] M [auth D] N [auth C] O [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | KY9100000034 |