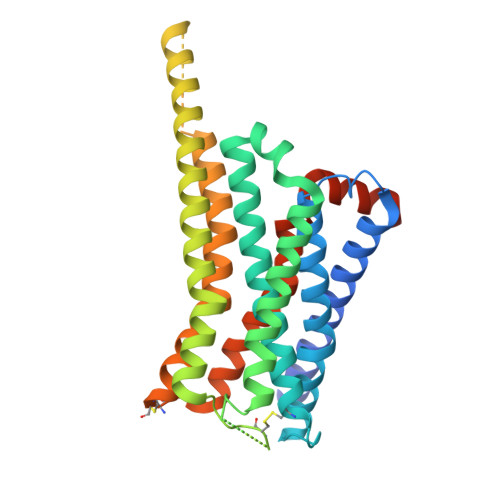
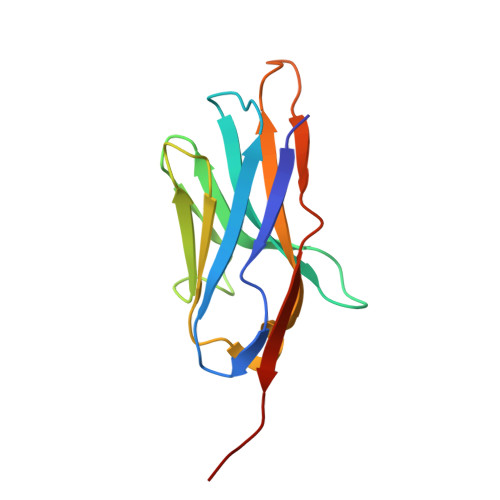
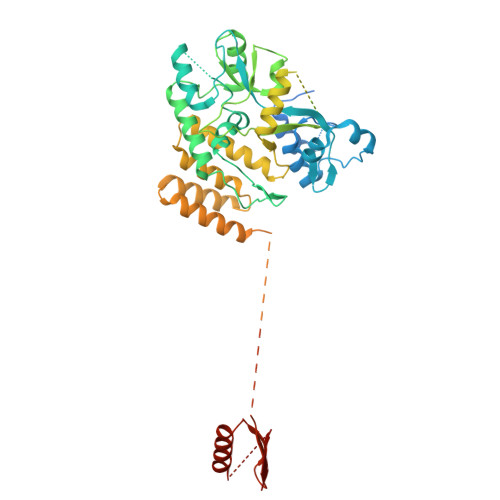
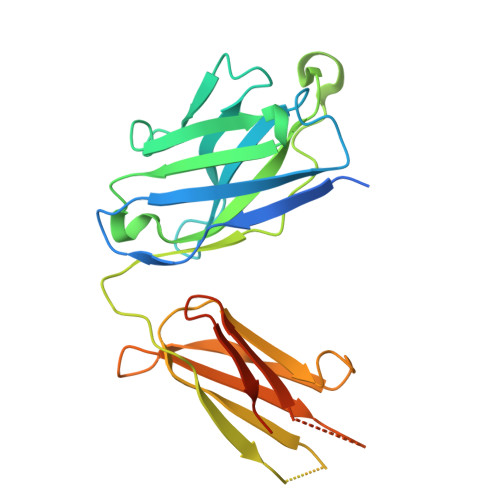
Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Fan, L., Zhuang, Y., Wu, H., Li, H., Xu, Y., Wang, Y., He, L., Wang, S., Chen, Z., Cheng, J., Xu, H.E., Wang, S.(2024) Neuron 112: 3295
- PubMed: 39094559
- DOI: https://doi.org/10.1016/j.neuron.2024.07.003
- Primary Citation of Related Structures:
8JXR, 8JXS - PubMed Abstract:
Understanding the kinetics of LSD in receptors and subsequent induced signaling is crucial for comprehending both the psychoactive and therapeutic effects of LSD. Despite extensive research on LSD's interactions with serotonin 2A and 2B receptors, its behavior on other targets, including dopamine receptors, has remained elusive. Here, we present cryo-EM structures of LSD/PF6142-bound dopamine D 1 receptor (DRD1)-legobody complexes, accompanied by a β-arrestin-mimicking nanobody, NBA3, shedding light on the determinants of G protein coupling versus β-arrestin coupling. Structural analysis unveils a distinctive binding mode of LSD in DRD1, particularly with the ergoline moiety oriented toward TM4. Kinetic investigations uncover an exceptionally rapid dissociation rate of LSD in DRD1, attributed to the flexibility of extracellular loop 2 (ECL2). Moreover, G protein can stabilize ECL2 conformation, leading to a significant slowdown in ligand's dissociation rate. These findings establish a solid foundation for further exploration of G protein-coupled receptor (GPCR) dynamics and their relevance to signal transduction.
- Key Laboratory of Systems Health Science of Zhejiang Province, School of Life Science, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou 310024, China. Electronic address: fanluyu2018@sibcb.ac.cn.
Organizational Affiliation: