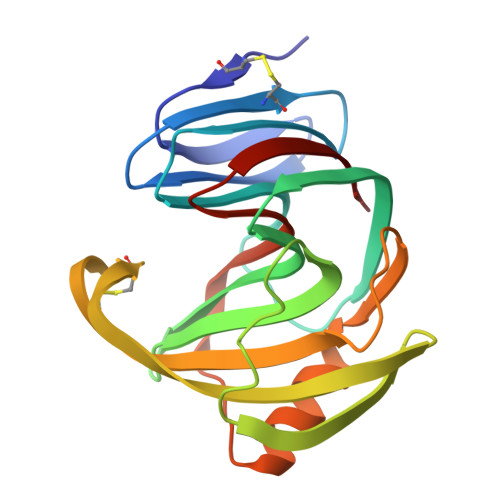
Disulfide bonds enhance thermal stability and thumb region drives activity of the glycoside hydrolase 11 xylanase rMxyl cd .
Wu, S., Zhang, N., Wan, Q.(2025) J Struct Biol 217: 108209-108209
- PubMed: 40368259
- DOI: https://doi.org/10.1016/j.jsb.2025.108209
- Primary Citation of Related Structures:
8JST - PubMed Abstract:
Thermostable enzymes have significant advantages in industries, yet uncovering novel candidates with superior properties remains a scientific pursuit. This study identified rMxyl cd , a glycoside hydrolase 11 family thermophilic xylanase from compost-soil metagenome, which exhibited a high specific activity of 5954 U·mg -1 at pH 5.5 and 80°C. rMxyl cd was crystallized and diffracted to 1.5 Å resolution. Compared to the mesophilic xylanase Xyn II, rMxyl cd exhibits a more compact architecture. Notably, B-factor analysis reveals a uniquely flexible thumb region, hinting at its critical role in the enzyme's catalytic mechanism. rMxyl cd contains two disulfide bonds in the thumb and the N-terminal regions. Breaking these disulfide bonds by mutagenesis has dramatically decreased activities and thermostability. Conversely, introducing an extra disulfide bond at the N-terminal region of its α-helix extended its half-life for more than five folds at 80°C. Our studies firmly establish that the disulfide bonds are essential for its high thermal stability and the flexibility of the thumb region is crucial for its activity. Comparing the rMxyl cd crystal structure with the AlphaFold2-predicted model shows overall similarity, but the crystal structure offers higher local accuracy, especially in key functional regions. These findings not only deepen our understanding of the structure-function relationship of thermophilic xylanases but also inform a rational design of industrial enzymes.
- Jiangsu Provincial Key Lab for Solid Organic Waste Utilization, Nanjing Agricultural University, Nanjing 210095, China; Key Lab of Organic-Based Fertilizers of China, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Collaborative Innovation Center for Solid Organic Wastes, Nanjing Agricultural University, Nanjing 210095, China; Educational Ministry Engineering Center of Resource-Saving Fertilizers, Nanjing Agricultural University, Nanjing 210095, China; College of Science, Nanjing Agricultural University, Nanjing 210095, China.
Organizational Affiliation: