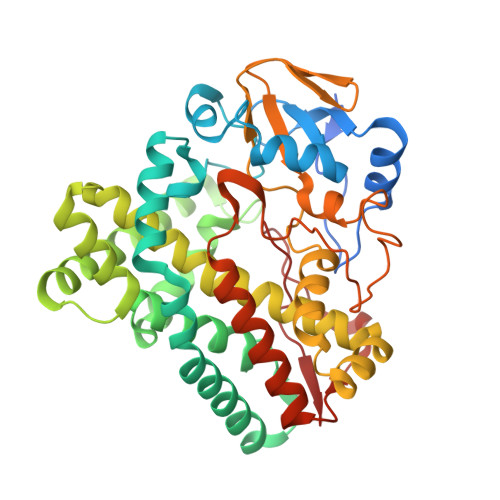
Regulation of P450 TleB catalytic flow for the synthesis of sulfur-containing indole alkaloids by substrate structure-directed strategy. and protein engineering.
Ge, X.Y., Long, Y., Wang, J., Gu, B., Yang, Z.X., Feng, Y.Y., Zheng, S., Li, Y.Y., Yan, W.P., Song, H.(2023) Sci China B Chem 66: 3232-3241