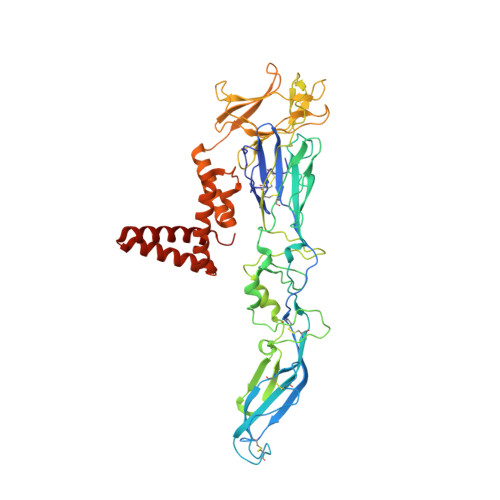
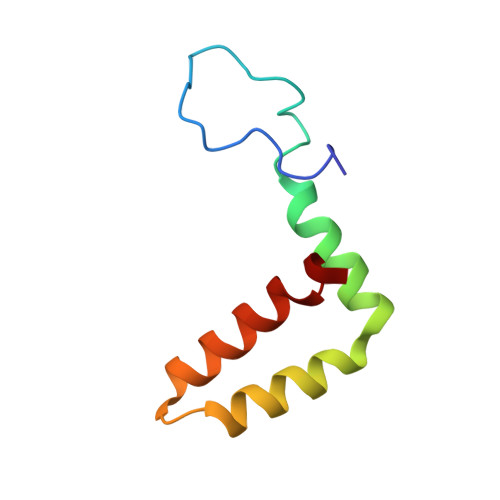
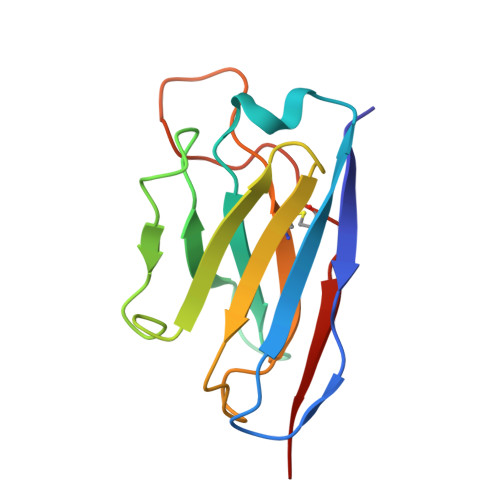
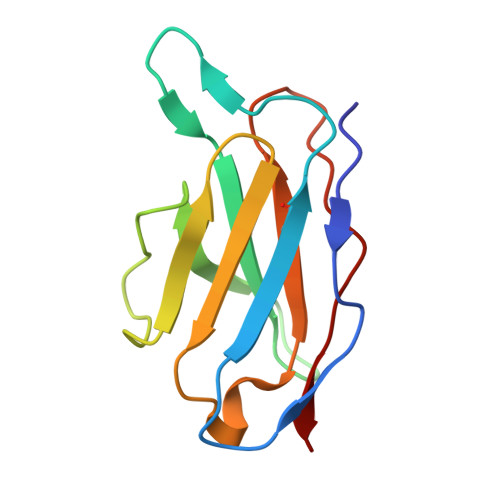
Ultrapotent human antibodies lock E protein dimers central region of diverse DENV3 morphological variants.
Fibriansah, G., Ng, T.S., Lim, X.N., Shebanova, A., Ng, L.C., Tan, S.L., Tan, A.W.K., Shi, J., Crowe Jr., J.E., Lok, S.M.(2025) Nat Commun 16: 9182-9182
- PubMed: 41102151
- DOI: https://doi.org/10.1038/s41467-025-64210-7
- Primary Citation of Related Structures:
8JN1, 8JN2, 8JN3, 8JN4, 8JN5, 8JN6, 8JN7 - PubMed Abstract:
Dengue virus (DENV) consists of four serotypes (DENV1-4). Current vaccines induce differing levels of immune response against the four serotypes, that might prime recipients to develop severe disease in subsequent infections. Several DENV tetravalent vaccine clinical trials suggested an increased incidence in severe DENV3 cases, suggesting a need to develop DENV3 therapeutics. Human monoclonal antibodies (HMAbs) DENV-290 and DENV-115 are ultrapotent against diverse DENV3 strains with differing particle morphologies. They mainly neutralize by inhibition of virus attachment to cells. CryoEM structures of Fabs complexed with differing DENV3 morphological variants show their Fabs binding across two E protein protomers at the center of the E dimer. This new class of E protein dimer binding antibodies is named EDE-C. The cryoEM structures also show how IgGs engage the DENV particles. Results define the structural and molecular basis for the ultrapotent activity of EDE-C antibodies.
- Programme in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore, Singapore.
Organizational Affiliation: