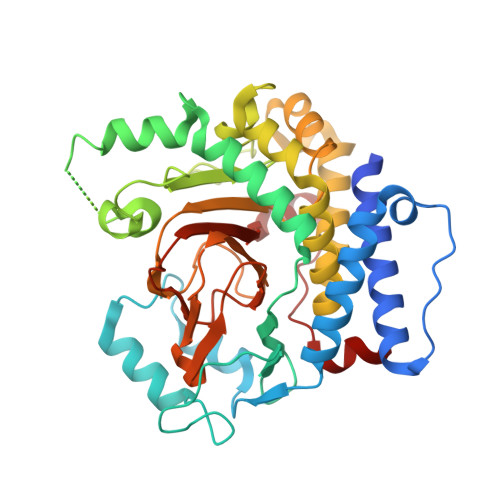
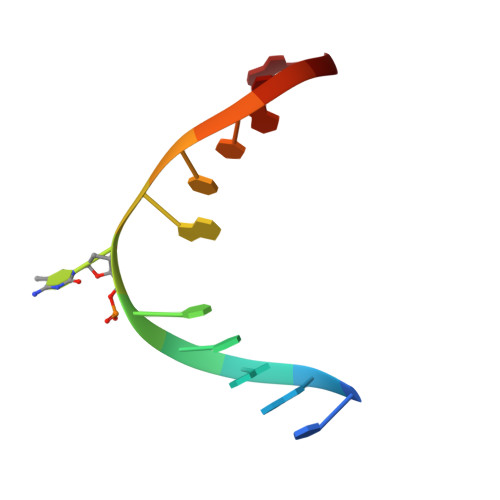
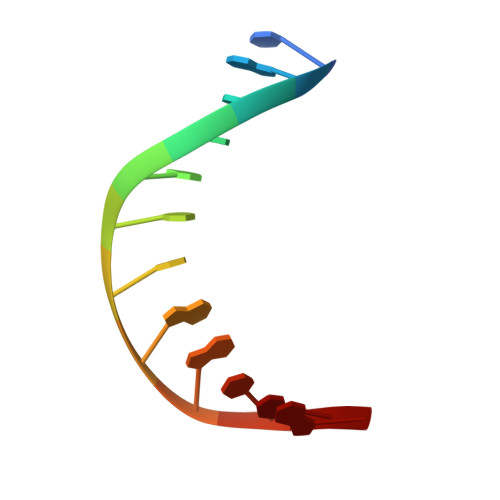
Molecular basis of an atypical dsDNA 5mC/6mA bifunctional dioxygenase CcTet from Coprinopsis cinerea in catalyzing dsDNA 5mC demethylation.
Zhang, L., Mu, Y., Li, T., Hu, J., Lin, H., Zhang, L.(2024) Nucleic Acids Res 52: 3886-3895
- PubMed: 38324471
- DOI: https://doi.org/10.1093/nar/gkae066
- Primary Citation of Related Structures:
8JKK - PubMed Abstract:
The eukaryotic epigenetic modifications 5-methyldeoxycytosine (5mC) and N6-methyldeoxyadenine (6mA) have indispensable regulatory roles in gene expression and embryonic development. We recently identified an atypical bifunctional dioxygenase CcTet from Coprinopsis cinerea that works on both 5mC and 6mA demethylation. The nonconserved residues Gly331 and Asp337 of CcTet facilitate 6mA accommodation, while D337F unexpectedly abolishes 5mC oxidation activity without interfering 6mA demethylation, indicating a prominent distinct but unclear 5mC oxidation mechanism to the conventional Tet enzymes. Here, we assessed the molecular mechanism of CcTet in catalyzing 5mC oxidation by representing the crystal structure of CcTet-5mC-dsDNA complex. We identified the distinct mechanism by which CcTet recognizes 5mC-dsDNA compared to 6mA-dsDNA substrate. Moreover, Asp337 was found to have a central role in compensating for the loss of a critical 5mC-stablizing H-bond observed in conventional Tet enzymes, and stabilizes 5mC and subsequent intermediates through an H-bond with the N4 atom of the substrates. These findings improve our understanding of Tet enzyme functions in the dsDNA 5mC and 6mA demethylation pathways, and provide useful information for future discovery of small molecular probes targeting Tet enzymes in DNA active demethylation processes.
- Department of Pharmacology and Chemical Biology, State Key Laboratory of Systems Medicine for Cancer, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Organizational Affiliation: