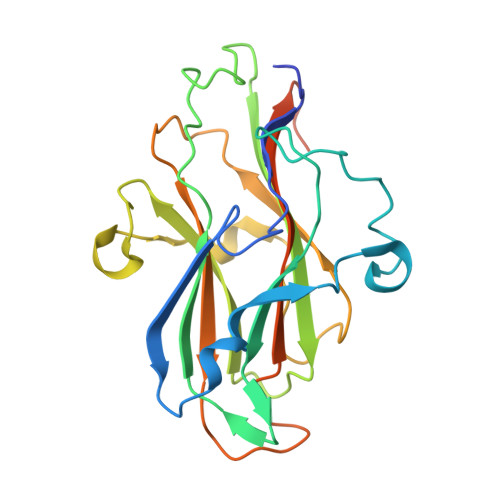
Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Liu, G., Chang, Y., Mei, X., Chen, G., Zhang, Y., Jiang, X., Tao, W., Xue, C.(2024) Int J Biol Macromol 255: 127959-127959
- PubMed: 37951443
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.127959
- Primary Citation of Related Structures:
8JIY - PubMed Abstract:
Chondroitin sulfate is a biologically and commercially important polysaccharide with a variety of applications. Carbohydrate-binding module (CBM) is an important class of carbohydrate-binding protein, which could be utilized as a promising tool for the applications of polysaccharides. In the present study, an unknown function domain was explored from a putative chondroitin sulfate lyase in PL29 family. Recombinant PhCBM100 demonstrated binding capacity to chondroitin sulfates with K a values of 2.1 ± 0.2 × 10 6 M -1 and 6.0 ± 0.1 × 10 6 M -1 to chondroitin sulfate A and chondroitin sulfate C, respectively. The 1.55 Å resolution X-ray crystal structure of PhCBM100 exhibited a β-sandwich fold formed by two antiparallel β-sheets. A binding groove in PhCBM100 interacting with chondroitin sulfate was subsequently identified, and the potential of PhCBM100 for visualization of chondroitin sulfate was evaluated. PhCBM100 is the first characterized chondroitin sulfate-specific CBM. The novelty of PhCBM100 proposed a new CBM family of CBM100.
- College of Food Science and Engineering, Ocean University of China, 1299 Sansha Road, Qingdao 266404, China.
Organizational Affiliation: