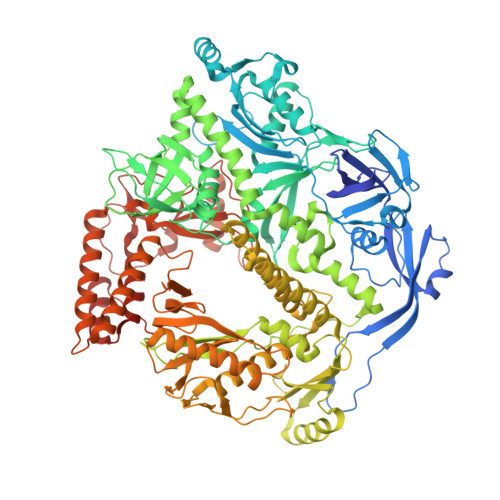
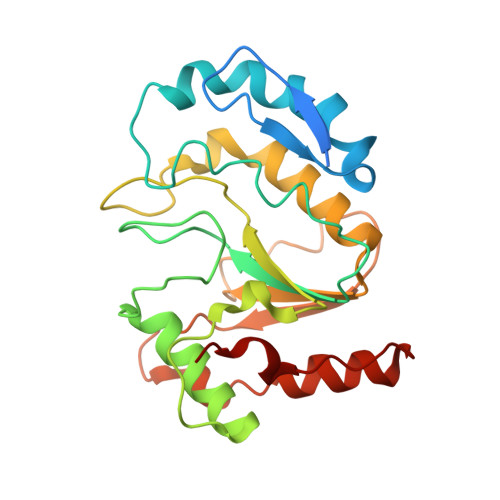
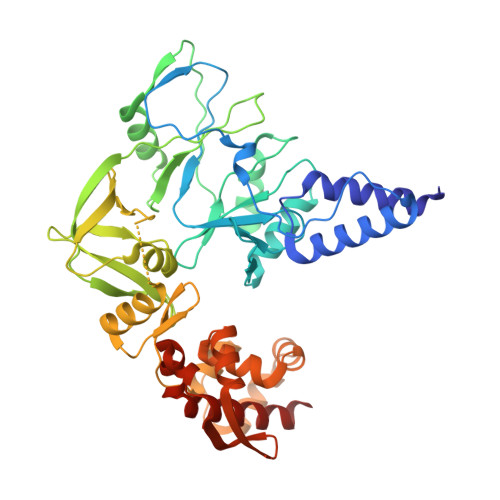
Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Xu, Y., Wu, Y., Wu, X., Zhang, Y., Yang, Y., Li, D., Yang, B., Gao, K., Zhang, Z., Dong, C.(2024) Int J Biol Macromol 270: 132231-132231
- PubMed: 38735603
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.132231
- Primary Citation of Related Structures:
8J86, 8J8F, 8J8G - PubMed Abstract:
Mpox virus has wildly spread over 108 non-endemic regions in the world since May 2022. DNA replication of mpox is performed by DNA polymerase machinery F8-A22-E4, which is known as a great drug target. Brincidofovir and cidofovir are reported to have broad-spectrum antiviral activity against poxviruses, including mpox virus in animal models. However, the molecular mechanism is not understood. Here we report cryogenic electron microscopy structures of mpox viral F8-A22-E4 in complex with a DNA duplex, or dCTP and the DNA duplex, or cidofovir diphosphate and the DNA duplex at resolution of 3.22, 2.98 and 2.79 Å, respectively. Our structural work and DNA replication inhibition assays reveal that cidofovir diphosphate is located at the dCTP binding position with a different conformation to compete with dCTP to incorporate into the DNA and inhibit DNA synthesis. Conformation of both F8-A22-E4 and DNA is changed from the pre-dNTP binding state to DNA synthesizing state after dCTP or cidofovir diphosphate is bound, suggesting a coupling mechanism. This work provides the structural basis of DNA synthesis inhibition by brincidofovir and cidofovir, providing a rational strategy for new therapeutical development for mpox virus and other pox viruses.
- Department of Thyroid and Breast Surgery, Zhongnan Hospital of Wuhan University, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery, State Key Laboratory of Virology, School of Pharmaceutical Sciences, Wuhan University, Wuhan 430071, China.
Organizational Affiliation: