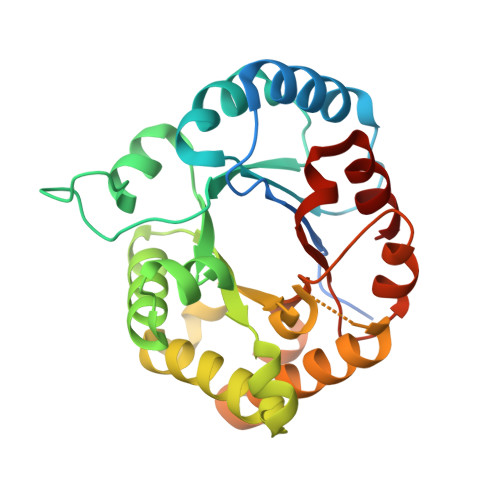
Crystal structure of Leishmania orientalis triosephosphate isomerase at 1.88 angstrom resolution and its specific inhibitors.
Kuaprasert, B., Leartsakulpanich, U., Riangrungroj, P., Pornthanakasem, W., Suginta, W., Mungthin, M., Leelayoova, S., Kiriwan, D., Choowongkomon, K.(2025) Biochimie 233: 27-35
- PubMed: 39984112
- DOI: https://doi.org/10.1016/j.biochi.2025.02.004
- Primary Citation of Related Structures:
8J7C - PubMed Abstract:
Leishmania orientalis, previously called L. siamensis, is a new species characterized as causing cutaneous leishmaniasis in Thailand. This study solves the crystal structure of the L. orientalis triosephosphate isomerase (LoTIM) in apo form at 1.88 Å resolution by using molecular replacement method. Tyrosine118 presents in the LoTIM protein sequence, whereas L. mexicana and Trypanosoma cruzi TIMs have a relative Cys118, which plays a major role in their specific ligand binding. Sulfur atom of the Cys57 thiol group is covalently bound to an arsenic (As) atom present in the precipitating solution. Although the electron density of loop-6 (Gly174-Tyr175-Gly176-Lys177-Val178) is missing in the structure due to this region lacking rigidity, the biological assembly of the two monomers of the LoTIM crystal structures are like that of L. mexicana and T. cruzi. 3D molecular protein-ligand docking was performed using the dimeric interfacial pocket of the enzyme as a ligand-binding receptor to identify its specific inhibitors. Five potential inhibiting compounds, including NSC639174, NSC606498, NSC110039, NSC58446, and NSC345647, were obtained with their IC 50 2.79 ± 0.10, 3.28 ± 0.80, 3.67 ± 0.11, 4.59 ± 0.87 and 15.44 ± 0.14 μM, respectively. However, specific inhibition assays against TIMs from L. orientalis and rabbit muscle indicate that NSC639174 and NSC110039 are the most potent inhibitors for LoTIM, whereas NSC58446 inhibits well both the parasitic and rabbit enzymes.
- Synchrotron Research and Applications Division, Synchrotron Light Research Institute (Public Organization), 111 University Avenue, Muang District, Nakhon Ratchasima, 30000, Thailand. Electronic address: buabarn@slri.or.th.
Organizational Affiliation: