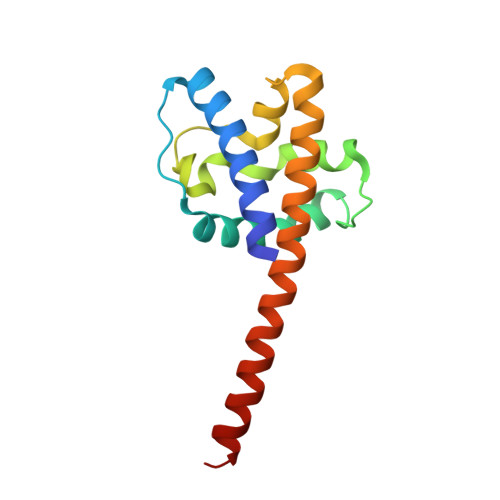
Structural analysis of conformational changes in the mpox virus A7 protein.
Ni, X., Wang, K., Han, Y., Lei, J.(2024) Virol Sin 39: 331-334
- PubMed: 38159644
- DOI: https://doi.org/10.1016/j.virs.2023.12.006
- Primary Citation of Related Structures:
8IZT, 8IZU - PubMed Abstract:
• Phospholipid-binding abilities of mpox virus A7 protein and its truncations are investigated. • The structures of the N-terminal truncations of A7 protein (A7N 121 and A7N 137 ) are determined. • Conformational changes of the conserved linking helix in A7 are illustrated. • A structural model of the full-length A7 protein is proposed.
- National Clinical Research Center for Geriatrics, and State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, 610041, China.
Organizational Affiliation: