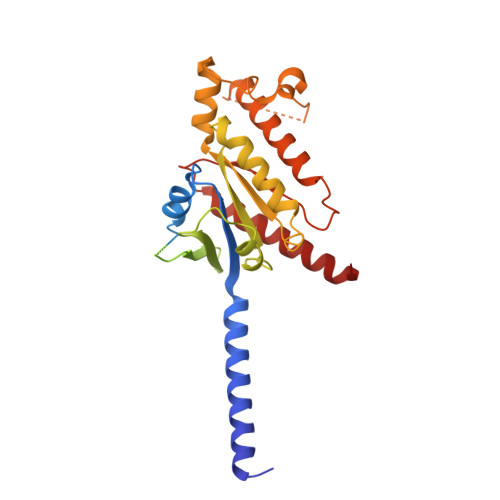
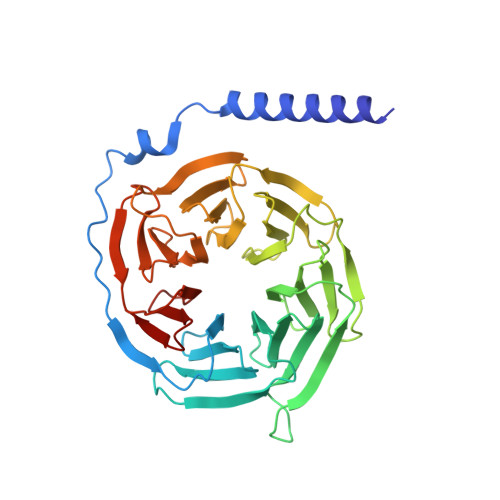
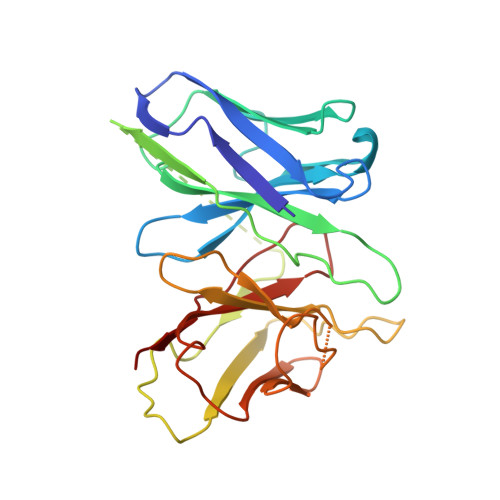
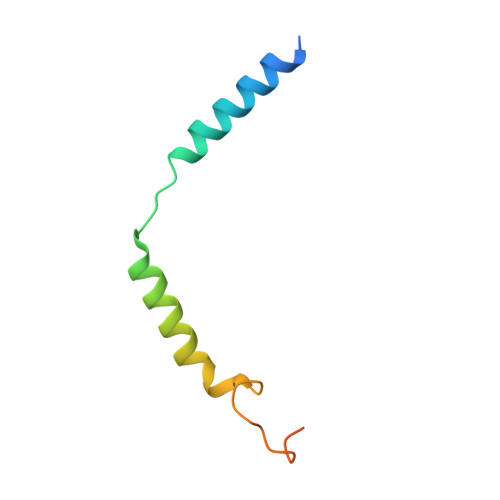
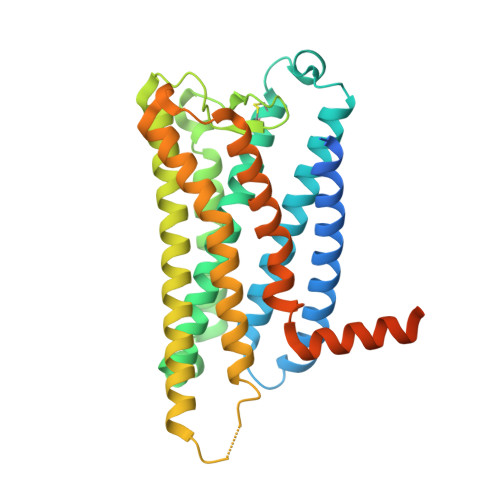
Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Wu, C., Xu, Y., He, Q., Li, D., Duan, J., Li, C., You, C., Chen, H., Fan, W., Jiang, Y., Eric Xu, H.(2023) Nat Commun 14: 2668-2668
- PubMed: 37160891
- DOI: https://doi.org/10.1038/s41467-023-38411-x
- Primary Citation of Related Structures:
8IUK, 8IUL, 8IUM - PubMed Abstract:
Prostaglandin F 2α (PGF 2α ), an endogenous arachidonic acid metabolite, regulates diverse physiological functions in many tissues and cell types through binding and activation of a G-protein-coupled receptor (GPCR), the PGF 2α receptor (FP), which also is the primary therapeutic target for glaucoma and several other diseases. Here, we report cryo-electron microscopy (cryo-EM) structures of the human FP bound to endogenous ligand PGF 2α and anti-glaucoma drugs LTPA and TFPA at global resolutions of 2.67 Å, 2.78 Å, and 3.14 Å. These structures reveal distinct features of FP within the lipid receptor family in terms of ligand binding selectivity, its receptor activation, and G protein coupling mechanisms, including activation in the absence of canonical PIF and ERY motifs and G q coupling through direct interactions with receptor transmembrane helix 1 and intracellular loop 1. Together with mutagenesis and functional studies, our structures reveal mechanisms of ligand recognition, receptor activation, and G protein coupling by FP, which could facilitate rational design of FP-targeting drugs.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China. wucanrong@simm.ac.cn.
Organizational Affiliation: