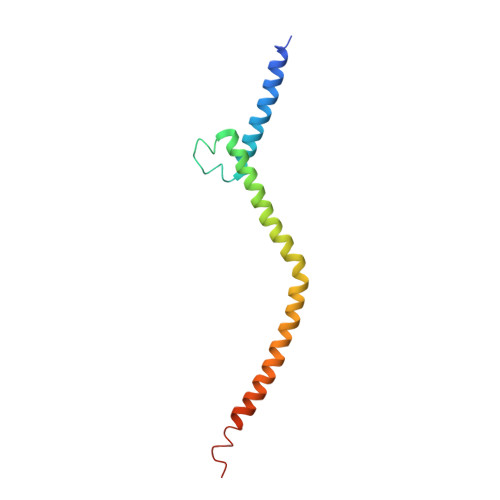
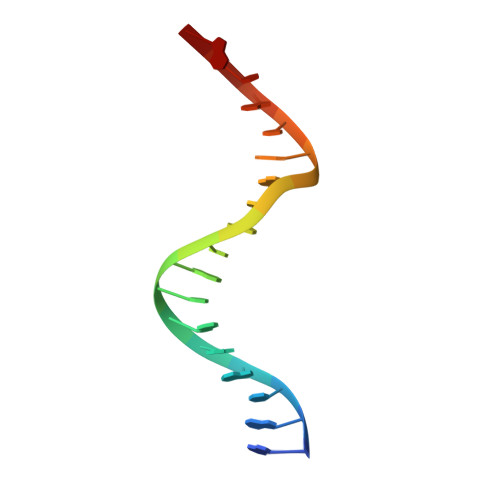
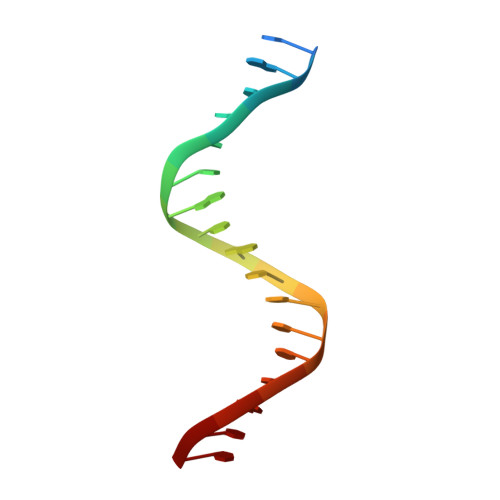
Tetramerization of upstream stimulating factor USF2 requires the elongated bent leucine zipper of the bHLH-LZ domain.
Huang, C., Xia, M., Qiao, H., Liu, Z., Lin, Y., Sun, H., Yu, B., Fang, P., Wang, J.(2023) J Biological Chem 299: 105240-105240
- PubMed: 37690682
- DOI: https://doi.org/10.1016/j.jbc.2023.105240
- Primary Citation of Related Structures:
8IA3 - PubMed Abstract:
Upstream stimulating factors (USFs), including USF1 and USF2, are key components of the transcription machinery that recruit coactivators and histone-modifying enzymes. Using the classic basic helix-loop-helix leucine zipper (bHLH-LZ) domain, USFs bind the E-box DNA and form tetramers that promote DNA looping for transcription initiation. The structural basis by which USFs tetramerize and bind DNA, however, remains unknown. Here, we report the crystal structure of the complete bHLH-LZ domain of USF2 in complex with E-box DNA. We observed that the leucine zipper (LZ) of USF2 is longer than that of other bHLH-LZ family transcription factors and that the C-terminus of USF2 forms an additional α-helix following the LZ region (denoted as LZ-Ext). We also found the elongated LZ-Ext facilitates compact tetramer formation. In addition to the classic interactions between the basic region and DNA, we show a highly conserved basic residue in the loop region, Lys271, participates in DNA interaction. Together, these findings suggest that USF2 forms a tetramer structure with a bent elongated LZ-Ext region, providing a molecular basis for its role as a key component of the transcription machinery.
- School of Materials and Chemistry, University of Shanghai for Science and Technology, Shanghai, China.
Organizational Affiliation: