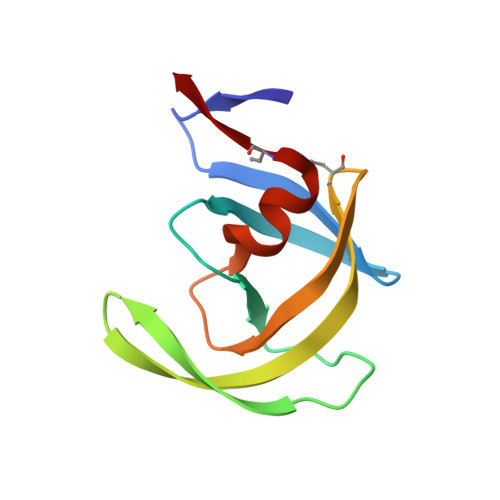
Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Jaskolski, M., Tomasselli, A.G., Sawyer, T.K., Staples, D.G., Heinrikson, R.L., Schneider, J., Kent, S.B., Wlodawer, A.(1991) Biochemistry 30: 1600-1609
- PubMed: 1993177
- DOI: https://doi.org/10.1021/bi00220a023
- Primary Citation of Related Structures:
8HVP - PubMed Abstract:
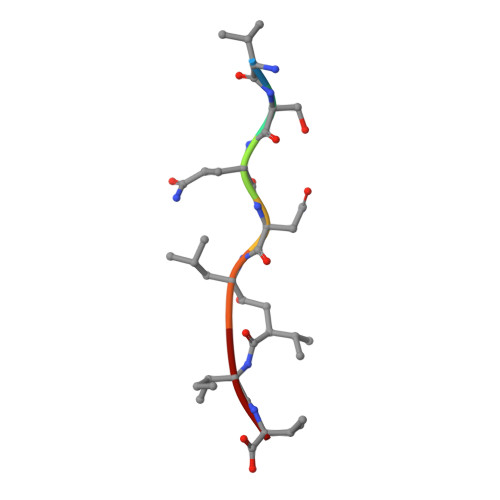
The crystal structure of a complex between chemically synthesized human immunodeficiency virus type 1 (HIV-1) protease and an octapeptide inhibitor has been refined to an R factor of 0.138 at 2.5-A resolution. The substrate-based inhibitor, H-Val-Ser-Gln-Asn-Leu psi [CH(OH)CH2]Val-Ile-Val-OH (U-85548e) contains a hydroxyethylene isostere replacement at the scissile bond that is believed to mimic the tetrahedral transition state of the proteolytic reaction. This potent inhibitor has Ki less than 1 nM and was developed as an active-site titrant of the HIV-1 protease. The inhibitor binds in an extended conformation and is involved in beta-sheet interactions with the active-site floor and flaps of the enzyme, which form the substrate/inhibitor cavity. The inhibitor diastereomer has the S configuration at the chiral carbon atom of the hydroxyethylene insert, and the hydroxyl group is within H-bonding distance of the two active-site carboxyl groups in the enzyme dimer. The two subunits of the enzyme are related by a pseudodyad, which superposes them at a 178 degrees rotation. The main difference between the subunits is in the beta turns of the flaps, which have different conformations in the two monomers. The inhibitor has a clear preferred orientation in the active site and the alternative conformation, if any, is a minor one (occupancy of less than 30%). A new model of the enzymatic mechanism is proposed in which the proteolytic reaction is viewed as a one-step process during which the nucleophile (water molecule) and electrophile (an acidic proton) attack the scissile bond in a concerted manner.
- Macromolecular Structure Laboratory, National Cancer Institute-Frederick Cancer Research and Development Center, Maryland 21702-1201.
Organizational Affiliation: