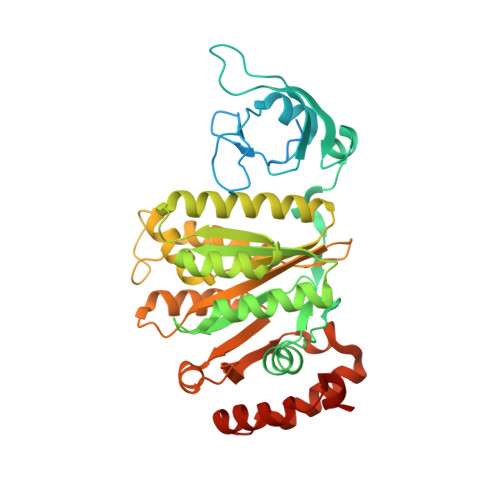
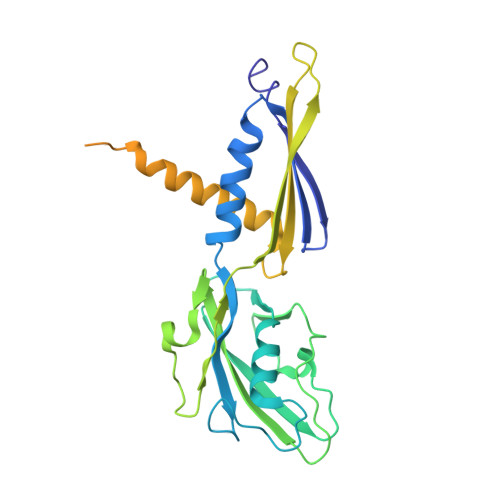
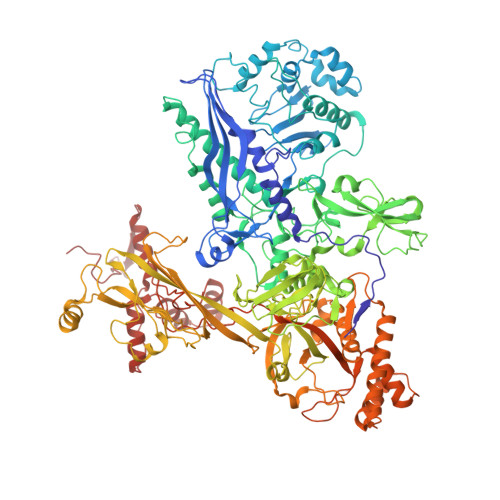
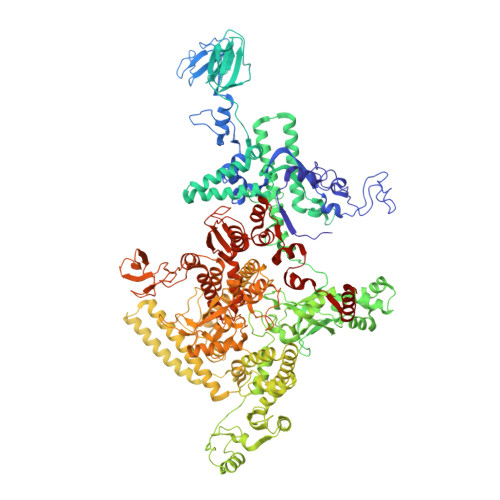
Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Murayama, Y., Ehara, H., Aoki, M., Goto, M., Yokoyama, T., Sekine, S.I.(2023) Sci Adv 9: eade7093-eade7093
- PubMed: 36753546
- DOI: https://doi.org/10.1126/sciadv.ade7093
- Primary Citation of Related Structures:
8HSG, 8HSH, 8HSJ, 8HSL, 8HSR - PubMed Abstract:
Transcription termination is an essential step in transcription by RNA polymerase (RNAP) and crucial for gene regulation. For many bacterial genes, transcription termination is mediated by the adenosine triphosphate-dependent RNA translocase/helicase Rho, which causes RNA/DNA dissociation from the RNAP elongation complex (EC). However, the structural basis of the interplay between Rho and RNAP remains obscure. Here, we report the cryo-electron microscopy structure of the Thermus thermophilus RNAP EC engaged with Rho. The Rho hexamer binds RNAP through the carboxyl-terminal domains, which surround the RNA exit site of RNAP, directing the nascent RNA seamlessly from the RNA exit to its central channel. The β-flap tip at the RNA exit is critical for the Rho-dependent RNA release, and its deletion causes an alternative Rho-RNAP binding mode, which is irrelevant to termination. The Rho binding site overlaps with the binding sites of other macromolecules, such as ribosomes, providing a general basis of gene regulation.
- Laboratory for Transcription Structural Biology, RIKEN Center for Biosystems Dynamics Research, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: