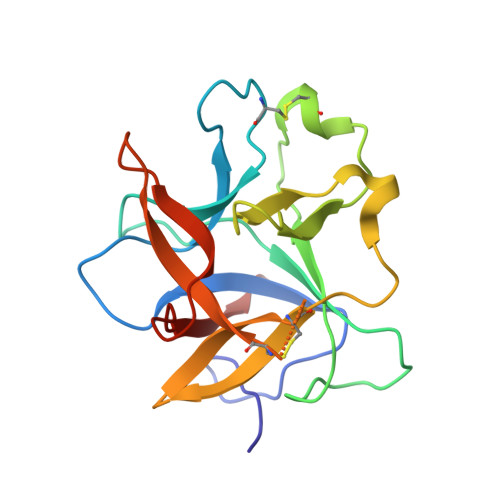
Crystal structure of Kunitz-type trypsin inhibitor: Entomotoxic effect of native and encapsulated protein targeting gut trypsin of Tribolium castaneum Herbst.
Mehmood, S., Thirup, S.S., Ahmed, S., Bashir, N., Saeed, A., Rafiq, M., Saeed, Q., Najam-Ul-Haq, M., Khaliq, B., Ibrahim, M., Alonazi, W.B., Akrem, A.(2024) Comput Struct Biotechnol J 23: 3132-3142
- PubMed: 39229336
- DOI: https://doi.org/10.1016/j.csbj.2024.07.023
- Primary Citation of Related Structures:
8HNR - PubMed Abstract:
Trypsin inhibitors are known to act against insect pests by inhibiting proteases of the digestive tract. In this study, we report structural and functional characterization of ∼ 19 kDa Albizia procera Kunitz-type trypsin inhibitor (ApKTI) protein with potential bio-insecticidal applications. Crystal structure of ApKTI protein has been refined to 1.42 Å and molecular structure (8HNR) showed highly beta sheeted conformation including 12 beta sheets, 15 loops and two small alpha helices. Docking between predicted model of Tribolium castaneum trypsin (TcPT) and 8HNR produced a stable complex (-11.3 kcal/mol) which reflects the inhibitory potential of ApKTI against insect gut trypsin. Significant mortality was observed in all life stages of T. castaneum including egg, larvae, pupae and adults with a 3.0 mg native ApKTI treatment in comparison to negative control. Although standard trypsin inhibitor ( Glycine max trypsin inhibitors; GmKTI; 3.0 mg) produced maximum reduction against all above life stages; however, a non-significant mortality difference was observed in comparison to 3.0 mg native ApKTI. The study further explores the synthesis and characterization of Graphene (GNPs) and Zinc oxide (ZnONPs) nanoparticles, followed by the optimization of ApKTI and GmKTI loading on both nanoparticles to evaluate their enhanced insecticidal effectiveness. Encapsulated proteins showed significant mortality against T. castaneum across all concentrations, with GNPs proving more effective than ZnONPs. Additionally, encapsulated GmKTI produced significant mortality of eggs compared to loaded ApKTI treatments while other life stages were non-significantly affected by two proteins. This research highlights the importance of encapsulated ApKTI protein for eco-friendly pest management strategies.
- Institute of Botany, Bahauddin Zakariya University, Multan 60800, Pakistan.
Organizational Affiliation: