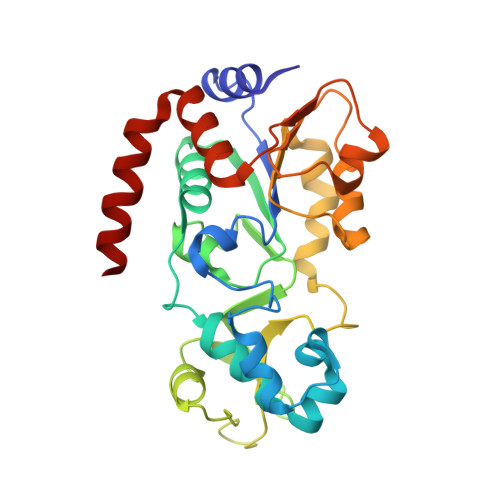
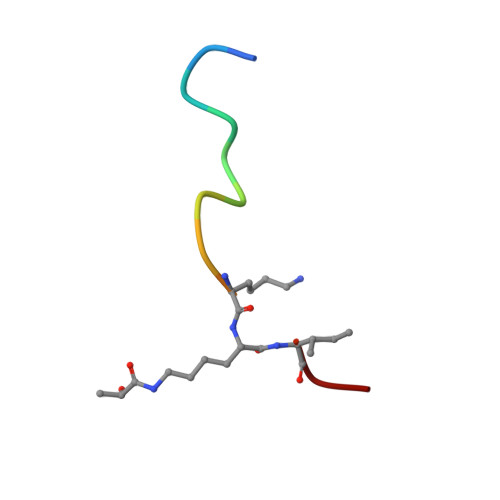
SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Jin, J., Bai, L., Wang, D., Ding, W., Cao, Z., Yan, P., Li, Y., Xi, L., Wang, Y., Zheng, X., Wei, H., Ding, C., Wang, Y.(2023) EMBO Rep 24: e56052-e56052
- PubMed: 36896611
- DOI: https://doi.org/10.15252/embr.202256052
- Primary Citation of Related Structures:
8HN9 - PubMed Abstract:
Lysine lactylation (Kla) is a recently discovered histone mark derived from metabolic lactate. The NAD + -dependent deacetylase SIRT3, which can also catalyze removal of the lactyl moiety from lysine, is expressed at low levels in hepatocellular carcinoma (HCC) and has been suggested to be an HCC tumor suppressor. Here we report that SIRT3 can delactylate non-histone proteins and suppress HCC development. Using SILAC-based quantitative proteomics, we identify cyclin E2 (CCNE2) as one of the lactylated substrates of SIRT3 in HCC cells. Furthermore, our crystallographic study elucidates the mechanism of CCNE2 K348la delactylation by SIRT3. Our results further suggest that lactylated CCNE2 promotes HCC cell growth, while SIRT3 activation by Honokiol induces HCC cell apoptosis and prevents HCC outgrowth in vivo by regulating Kla levels of CCNE2. Together, our results establish a physiological function of SIRT3 as a delactylase that is important for suppressing HCC, and our structural data could be useful for the future design of activators.
- Division of Life Sciences and Medicine, Department of Hepatobiliary Surgery, The First Affiliated Hospital of USTC, University of Science and Technology of China, Heifei, China.
Organizational Affiliation: