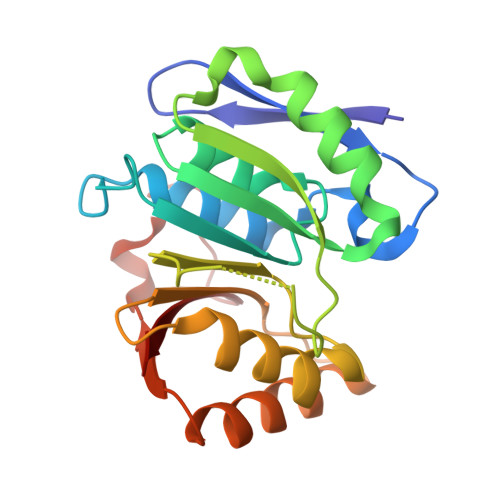
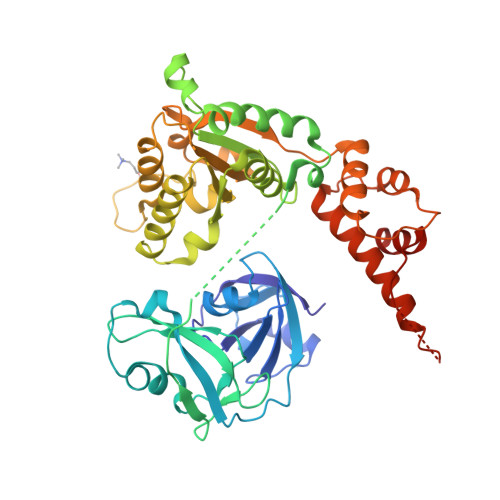
Structural basis for recognition and methylation of p97 by METTL21D, a valosin-containing protein lysine methyltransferase.
Nguyen, T.Q., Koh, S., Kwon, J., Jang, S., Kang, W., Yang, J.K.(2023) iScience 26: 107222-107222
- PubMed: 37456834
- DOI: https://doi.org/10.1016/j.isci.2023.107222
- Primary Citation of Related Structures:
8HL6, 8HL7 - PubMed Abstract:
p97 is a human AAA+ (ATPase associated with diverse cellular activities, also known as valosin-containing protein [VCP]) ATPase, which is involved in diverse cellular processes such as membrane fusion and proteolysis. Lysine-specific methyltransferase of p97 (METTL21D) was identified as a class I methyltransferase that catalyzes the trimethylation of Lys315 of p97, a so-called VCP lysine methyltransferase (VCPKMT). Interestingly, VCPKMT disassembles a single hexamer ring consisting of p97-D1 domain and methylates Lys315 residue. Herein, the structures of S-adenosyl-L-methionine-bound VCPKMT and S-adenosyl-L-homocysteine-bound VCPKMT in complex with p97 N/D1 (N21-Q458) were reported at a resolution of 1.8 Å and 2.8 Å, respectively. The structures revealed the molecular details for the recognition and methylation of monomeric p97 by VCPKMT. Using biochemical analysis, we also investigated whether the methylation of full-length p97 could be sufficiently enhanced through cooperation between VCPKMT and the C terminus of alveolar soft part sarcoma locus (ASPL). Our study provides the groundwork for future structural and mechanistic studies of p97 and inhibitors.
- Department of Chemistry and Integrative Institute of Basic Science, College of Natural Sciences, Soongsil University, Seoul 06978, Republic of Korea.
Organizational Affiliation: